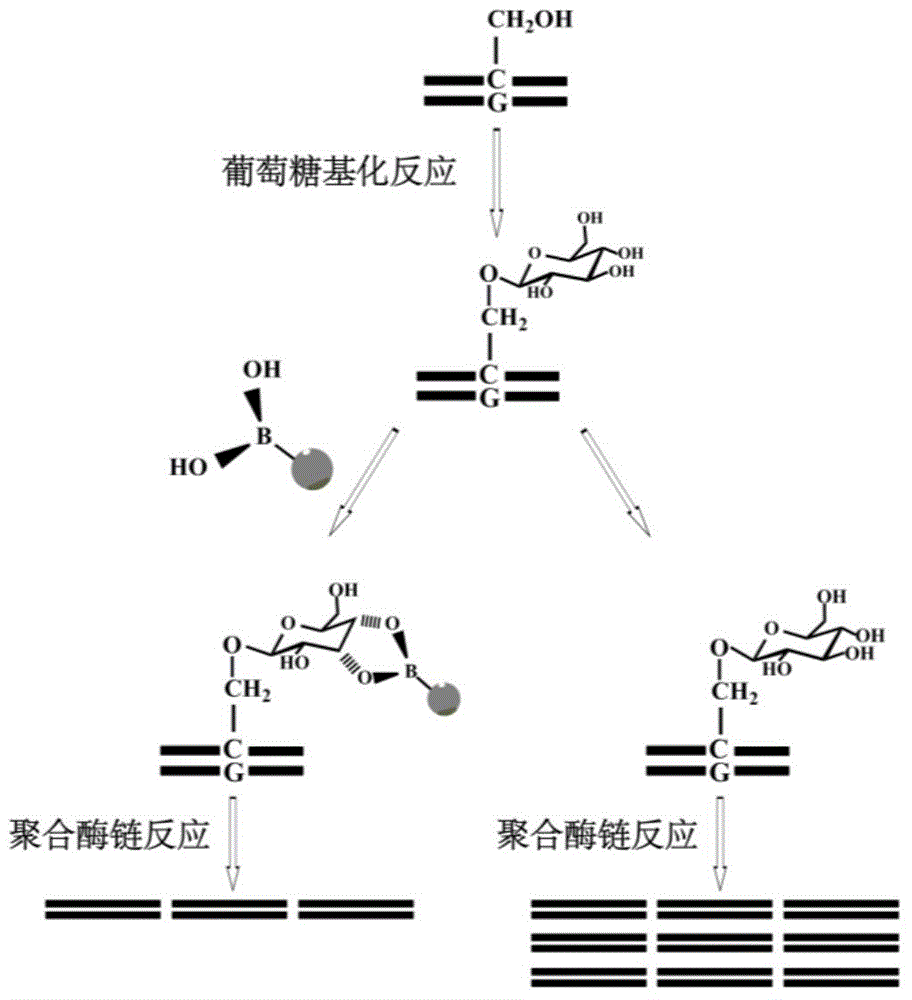
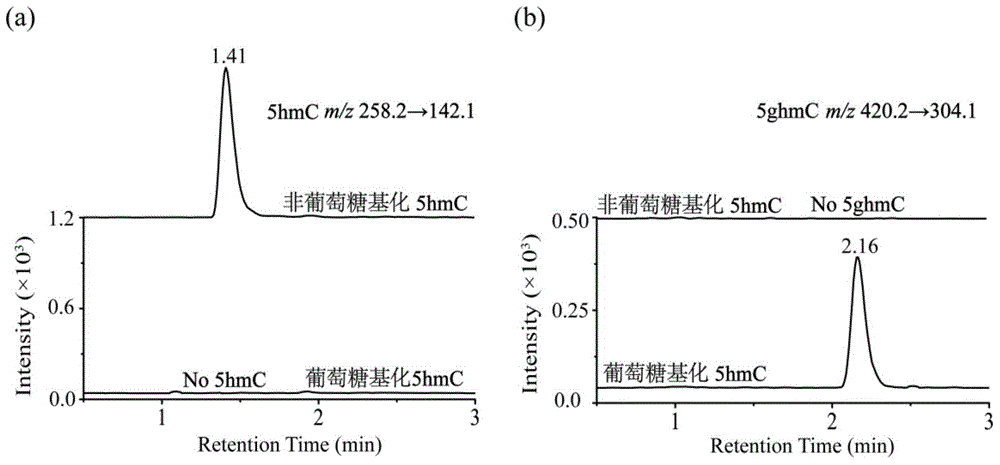
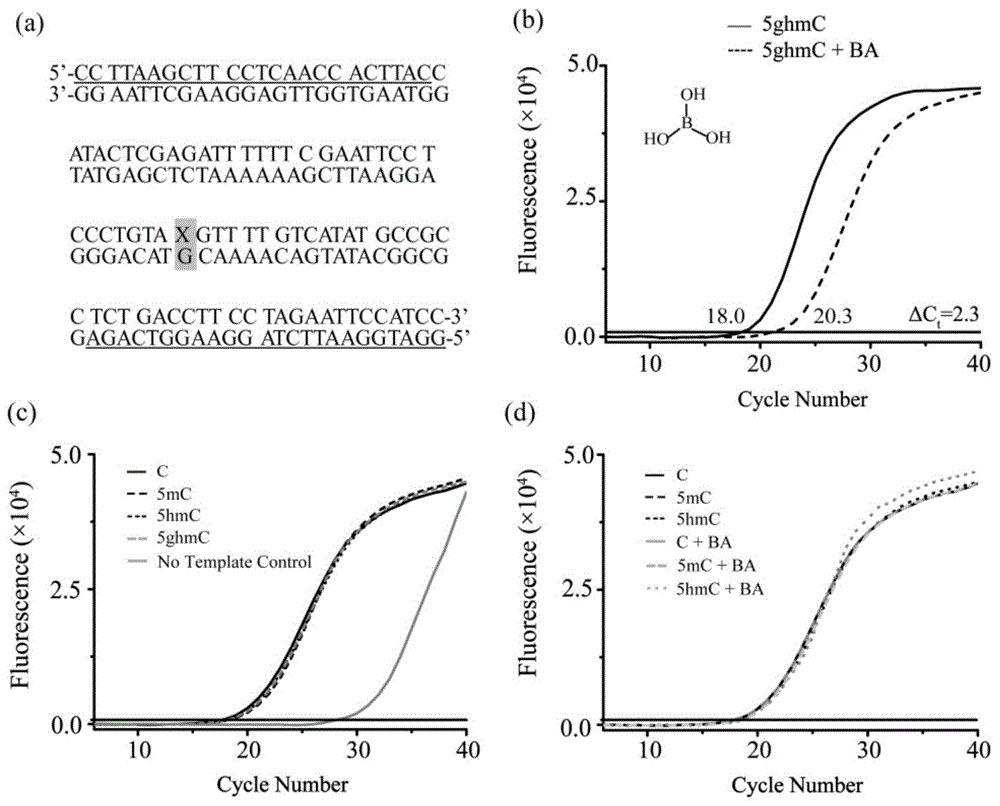
Method and kit for detecting 5-hydroxymethyl cytosine in DNA through boric acid mediated polymerase chain reactions
A technology of hydroxymethylcytosine and a kit, applied in the field of biological detection, can solve problems such as inability to distinguish gene sequences
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1. Using boric acid-mediated polymerase chain reaction kit to detect 5hmC of specific gene sequence of genomic DNA
[0041] The present invention uses a boric acid-mediated polymerase chain reaction kit to analyze the genomic DNA of mouse embryonic stem cells rich in 5hmC (5hmC content is 800 pieces / 10 6 C) The specific gene sequence 5hmC was identified and analyzed.
[0042] A highly enriched gene region of 5hmC was detected using this kit. In the results of mouse embryonic stem cell genomic DNA high-throughput sequencing (Illumina ChIP-Seq, GSE43262), three PCR amplification regions (INTRON_PAX5_1, INTRON_PAX5_2, INTRON_PAX5_3) of the B cell transcription factor related gene PAX5 were selected, and UTR-5_SRR was selected as the Quantitative PCR internal reference genes, design specific primers for different gene sequences, and then sequentially perform glucosylation and 2-CB-PBA treatment on the genomic DNA, and use the designed specific primers for PCR amplificati...
Embodiment 2
[0044] Example 2.5hmC quantitative analysis
[0045] Taking the sample treated with 2-CB-PBA as an example, the negative control probe (5hmC-ds100mer) and the positive control probe (5ghmC-ds100mer) are mixed in different proportions, and the molar concentration ratios of 5ghmC: 5hmC are selected as: 0:1, 1:16, 1:8, 1:4, 1:2, 1:1, 2:1, 4:1, and 1:0, and the mixed template concentration is maintained at 5.0nM. Then use this mixture as a PCR amplification template to investigate the 5ghmC-ds100mer concentration and ΔC t The relationship between the values so that the curve C can be amplified by PCR t The value change is used to quantitatively analyze 5ghmC. Attached Figure 5 a and Figure 5 As shown in b, as the concentration of 5ghmC increases, ΔC t The value increases from 0 to 6.0, and the linear relationship between the two is satisfied: ΔC t =6.14+4.87log[5ghmC / ds100mer](R 2 =0.97). Although ΔC t Value increases, but the amount of PCR amplification product is gradually dec...
Embodiment 3
[0046] Example 3. Comparison of boric acid-mediated polymerase chain reaction kit and restriction endonuclease-binding PCR detection kit (ie MspI enzymatic hydrolysis)
[0047] The restriction endonuclease MspI recognizes the CCGG sequence, and its digestion specificity is changed by glucosylation: MspI recognizes and cleaves 5mC and 5hmC, but cannot cleave 5ghmC; when the 5ghmC site is on the CCGG sequence, it can The cleaved 5hmC site was converted into a non-cleavable 5ghmC site, so that the sequence was preserved by PCR amplification. Therefore, the restriction endonuclease method is restricted by various conditions such as restriction sites.
[0048] First, use the positive control probe (5ghmC-ds100mer) as the PCR amplification template to investigate the PCR amplification conditions after 2-CB-PBA treatment and MspI digestion. After the probe is digested by MspI, C t The value does not change significantly, that is, the enzymatic hydrolysis treatment does not affect the PCR...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com