Novel exonuclease-III-based mercury ion detection method
A technology of exonuclease and mercury ions, which is applied in the field of analytical chemistry technology and food safety, and can solve the problems of expensive equipment and complicated pretreatment
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1 Design and synthesis of corresponding oligonucleotide fragments
[0025] By consulting relevant literature, design two pieces of DNA fragments that can specifically recognize mercury ions and are rich in T bases, and design upstream and downstream primers for real-time fluorescent quantitative PCR based on the sequences. Sequences were prepared by a DNA synthesizer.
[0026] Template DNA: 5'- AATCTGGTTTAGCTACGCCTTCCCCGTGGCGATGTTTCTT
[0027] AGCGCCTTACTTGTTTGTTG-3' (SEQ ID NO: 1)
[0028] Complementary DNA: 5'-CTTCTTTCTTGTAAGGCGCTAAGAAACATCGCCACGGG
[0029] GAAGGCGTACTCGCG-3' (SEQ ID NO: 2)
[0030] Upstream primer: 5'-AATCTGGTTTAGCTACGCCTTC-3' (SEQ ID NO: 3)
[0031] Downstream primer: 5'-GTAAGGCGCTAAGAAACATCG-3' (SEQ ID NO: 4)
Embodiment 2
[0032] Example 2 T-Hg 2+ -T hybridization reaction
[0033] First, add the mixture of template DNA and complementary DNA to the PCR tube, and then add an appropriate amount of Hg 2+ The ion standard solutions were made to have final concentrations of 2, 5, 10, 20, 50, 100, and 200 nM, respectively, and reacted for 30 min. Template DNA and Complementary DNA by T-Hg 2+ The -T-specific structure forms double-stranded DNA with a closed 3' end.
Embodiment 3
[0034] Enzyme catalysis of embodiment 3 exonuclease III and establishment of standard curve
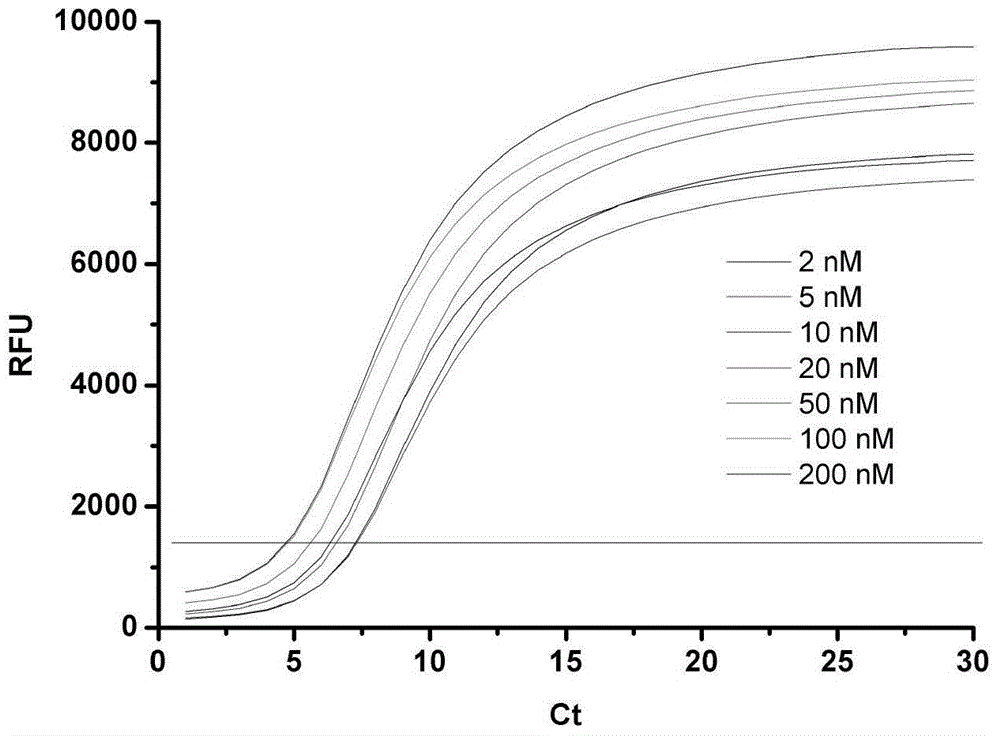
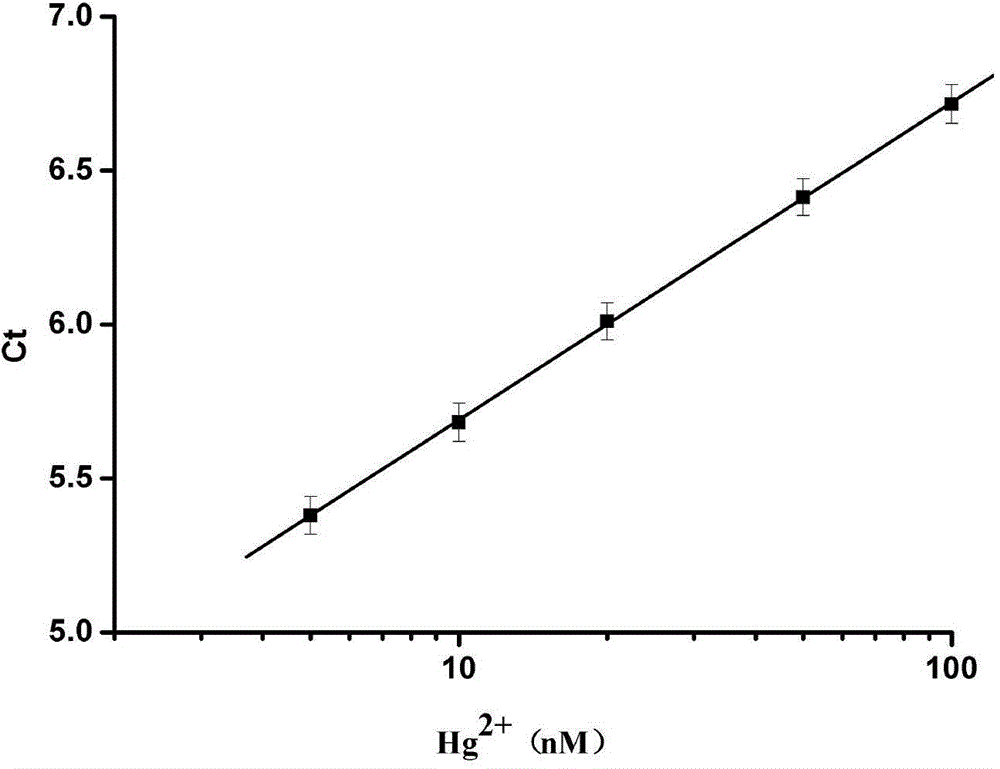
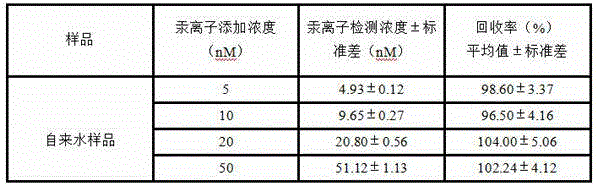
[0035] Add 0.1 μL of exonuclease III to the above-mentioned PCR tubes in turn, react for 90 s, and then react the whole system in a constant temperature metal bath at 98°C for 10 min to inactivate exonuclease III and end the reaction of exonuclease III. Shearing of template DNA. Then add an appropriate amount of PCR mixture, after the upstream and downstream primers in CFX96 TM Experiments were completed in real-time fluorescent quantitative PCR. Due to the added Hg 2+ Different ion concentrations lead to different amounts of template DNA being cut off by exonuclease III, and the concentration of template DNA used for PCR amplification is also different, so different amplification curves are obtained ( figure 1 ). The establishment of the mercury ion standard curve: Utilize the real-time fluorescent quantitative PCR instrument to measure the number of cycles of the amplification c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com