Dual fluorescent quantitative PCR method for species identification of salmons and highly processed products of salmons
A dual fluorescence and product variety technology, applied in the field of dual fluorescence quantitative PCR detection, can solve the problems of low cost, messy sequencing results, and inapplicability of mixed sample identification, and achieve the effect of shortening the detection time and reducing the cost of reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
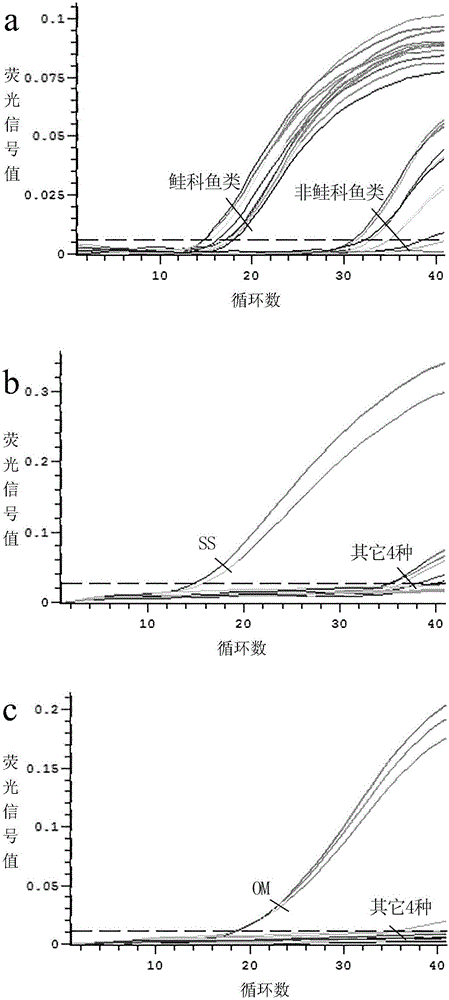
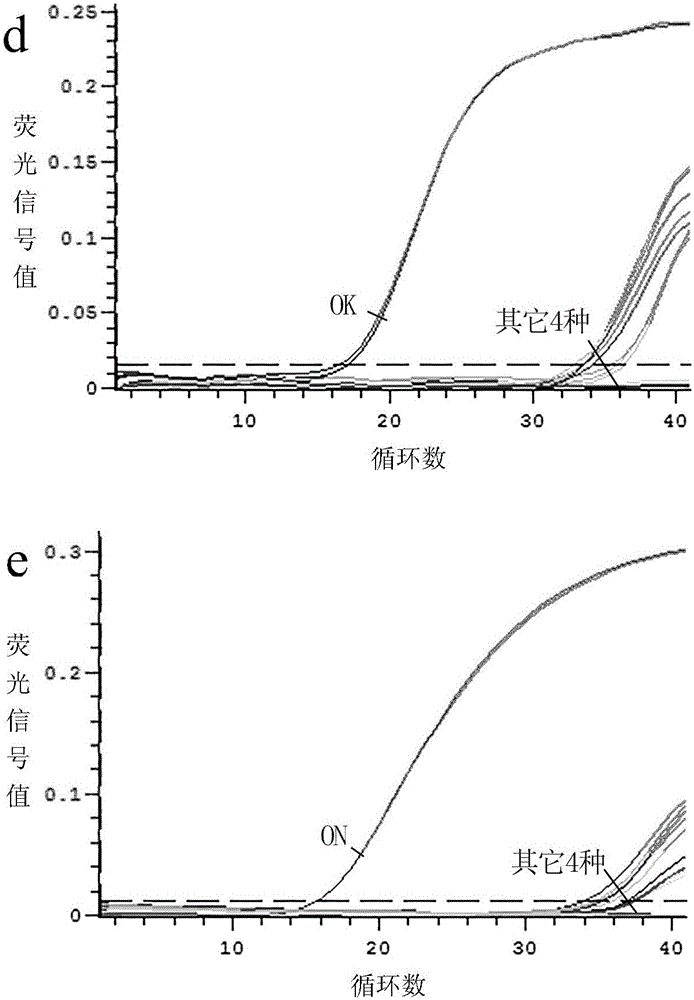
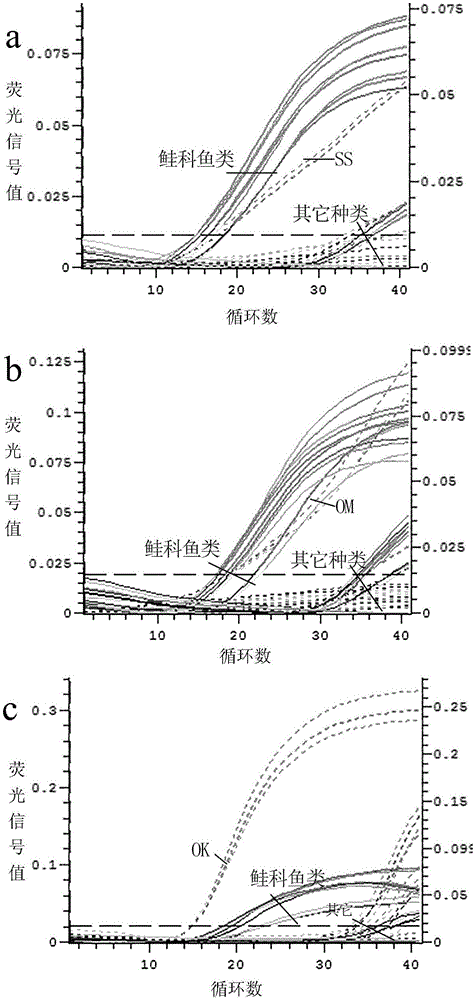
[0090] Embodiment 1, adopt above-mentioned double fluorescence quantitative PCR detection method to carry out species identification to the salmon sashimi that buys randomly on the market:
[0091] 1) Design of primers and probes for specific identification of Salmonidae and 4 common salmon species;
[0092] Specifically as described in Table 1.
[0093] 2), the extraction of genomic DNA in the salmon sample to be tested;
[0094] The sashimi marked as salmon was randomly purchased from the market, and the sample DNA was extracted with the Axygen Genomic DNA Mini-extraction Kit, and stored at -20°C.
[0095] 3) Fluorescence quantitative PCR amplification system setting:
[0096] The species were identified by double fluorescent quantitative PCR.
[0097] The dual fluorescent quantitative PCR system is: AceQ is contained in a total volume of 20 μL TM qPCRProbeMasterMix 10 μL, 0.4 μL each of salmon-specific upstream and downstream primers (5 μM), 0.2 μL of salmon-specific pr...
Embodiment 2
[0111] Embodiment 2, using the above-mentioned double fluorescent quantitative PCR detection method to carry out the species identification of the salmon and fish floss commodities purchased randomly on the market:
[0112] Randomly purchased goods marked as salmon fish floss from the market were used to extract sample DNA using Tiangen Deep Processed Food DNA Extraction Kit (DP326), and stored at -20°C. The species were identified by double fluorescent quantitative PCR. If the 16S rDNA detection gene of Salmonidae has no S-type amplification curve and / or the Ct value is greater than 30, the judgment result is negative, and it can be determined that the fish floss sample does not contain salmonid fish components; if the Ct value is less than 30, it can be judged as negative. If the result of the sample is determined to be positive, it is determined that the sample contains components of salmonids. If the CR detection genes of the 4 common salmon species have no S-curve genera...
Embodiment 3
[0119] Embodiment 3, using the above-mentioned double fluorescent quantitative PCR detection method to carry out the species identification of the canned salmon fish goods purchased randomly on the market:
[0120] Randomly buy canned salmon products from the market, use Tiangen Deep Processed Food DNA Extraction Kit (DP326) to extract DNA from samples, and store them at -20°C. The species were identified by double fluorescent quantitative PCR. If the 16S rDNA detection gene of Salmonidae has no S-type amplification curve and / or the Ct value is greater than 30, the judgment result is negative, and it can be determined that the canned fish sample does not contain salmonid fish components; if the Ct value is less than 30, it can be determined If the result of the sample is determined to be positive, it is determined that the sample contains components of salmonids. If the CR detection genes of the 4 common salmon species have no S-curve generation and / or the Ct value is greater...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com