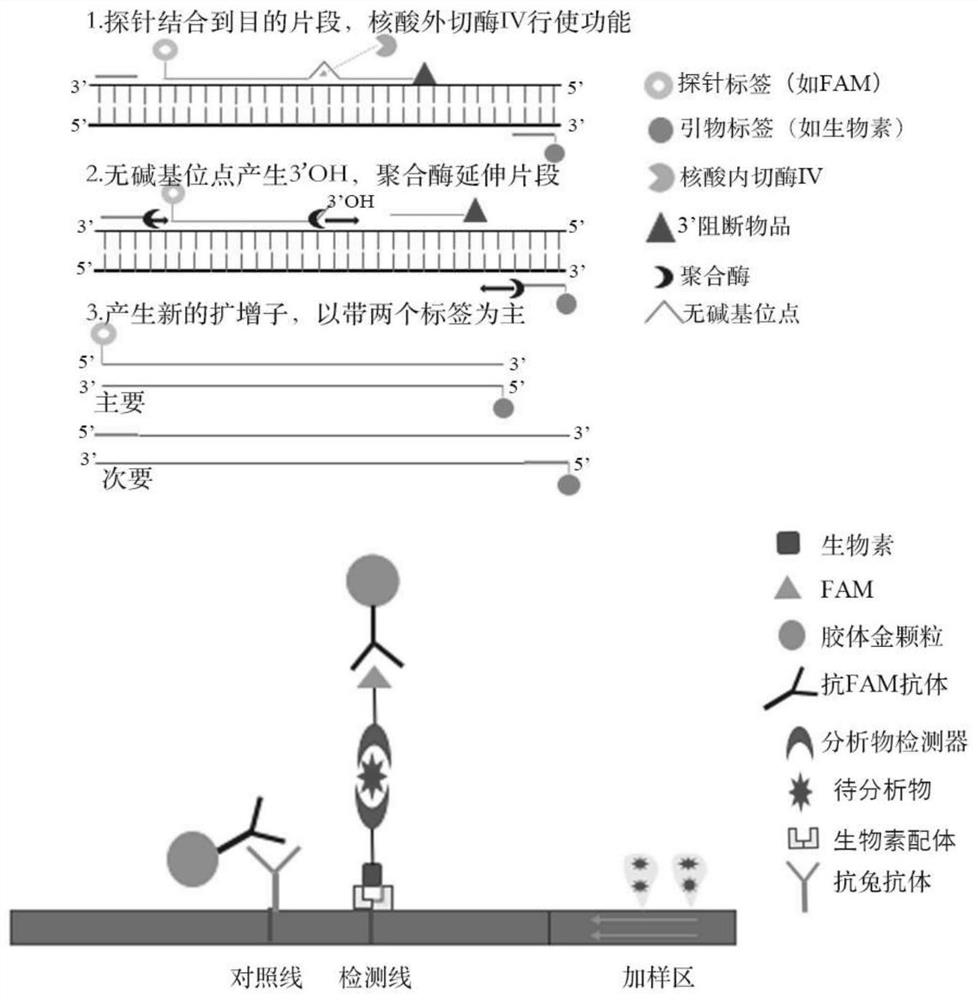
Novel coronavirus RPA test strip detection kit
A technology for detection kits and kits, which is applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of no design standards, low success rate, and difficulty in manual design, etc., to reduce reagents Cost, detection time saving, high sensitivity effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0034] Embodiment 1 RPA kit (test strip method) composition and method of use
[0035] The main components of the kit are shown in Table 1.
[0036] Table 1 The main components of the kit
[0037]
[0038]
[0039] Note: Packaging specification: 48 times / box bag, you need to prepare your own virus nucleic acid extraction kit, RNase inhibitor (recommended: Takara: article number: 9766, 2313Q); nucleic acid detection test strip: composed of sample pad, cellulose film, Consisting of detection belt, control belt and absorbent pad, it is recommended to use Universal nucleic acid detection test strip or disposable nucleic acid detection device (model: D001-3) of Hangzhou Ustar Biotechnology Co., Ltd.
[0040] The detailed introduction of primers, probes and other reagents in Table 1 is as follows:
[0041] (1) Primers, probes
[0042] The primer and probe sequences of the present invention are shown in Table 1. In the probe sequence, the 5' is modified by the fluorescent ...
experiment example 1
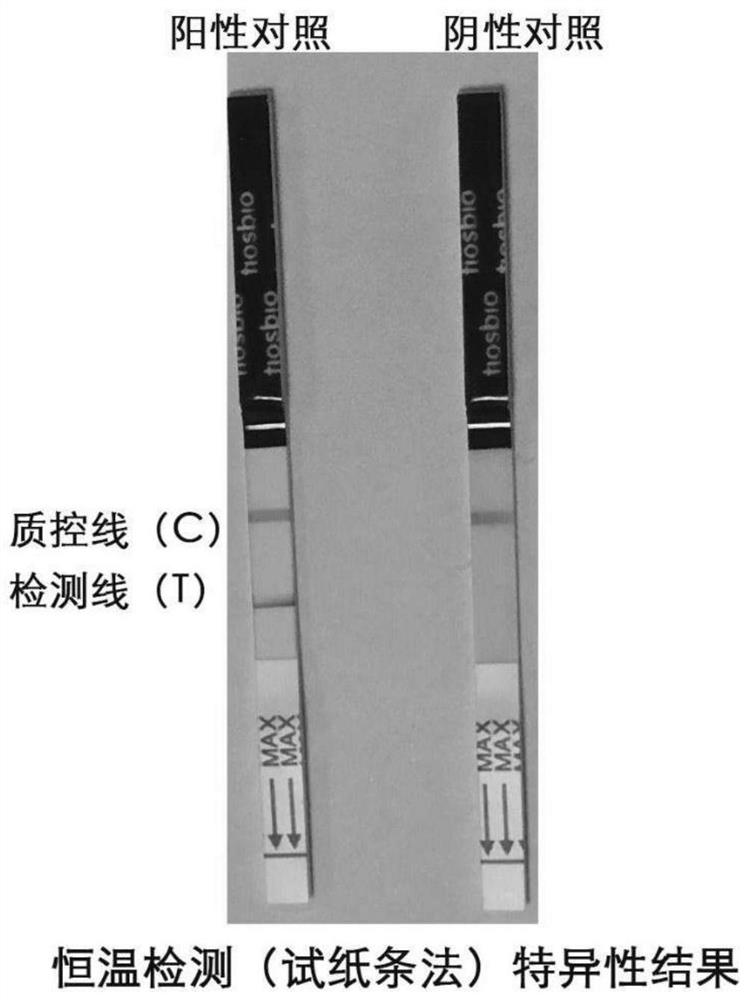
[0059] Experimental example 1 Specificity and sensitivity detection of RPA kit for detection of novel coronavirus
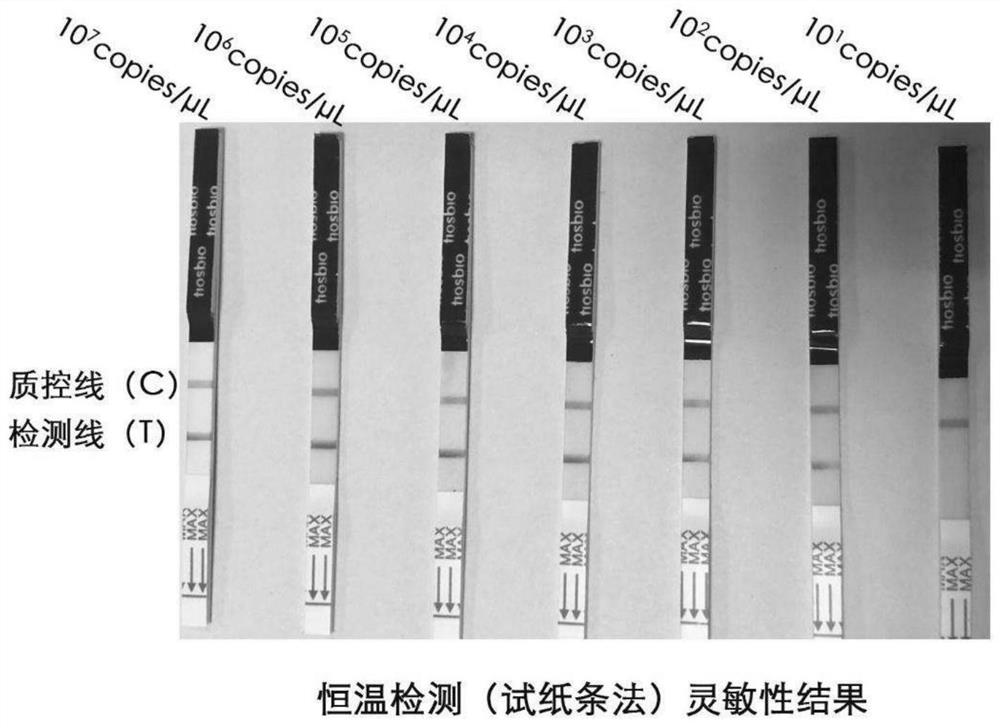
[0060] 1. Sensitivity detection:
[0061] The concentration of the plasmid standard was 40ng / μL measured by a UV spectrophotometer, the base number of pUC57-KANA was 2710bp, the target fragment size was 253bp, and the average relative molecular mass of each base was 660 Daltons / bp. The copy number in is calculated according to the formula: sample copy number (copies / ul) = (6.02×10 23 )×plasmid concentration (ng / μL)×10 -9 / (660×base number), the final calculated copy number is 1.23×10 10 copies / ul. The above standard is diluted according to 10-fold ratio, and the concentration is taken at 10 7 -10 1 Copies / ul was used for the experiment, reacted at 39°C for 10 minutes, after the reaction was completed, 5 μL of the reaction liquid was drawn, diluted 10-50 times by adding 1×PBST, and put into a test strip for 5-20 minutes.
[0062] The result is as figure 2...
experiment example 2
[0066] Experimental Example 2 Primer Design
[0067] At present, there is no definite RPA primer sequence design rule, and no design software is available. The inventor's primer design mainly follows the following rules:
[0068] 1) Primers should be 30-35 bases in length for optimal formation of recombinase / primer filaments; longer primers (>45 bases) are not recommended;
[0069] 2) Long tracks of a specific nucleotide or a large number of small repeats (such as AAAAA) should be avoided;
[0070] 3) RPA can amplify long sequences up to 1.5kb, but better results can be obtained with shorter amplicons in the range of 80–400bp (optimally 100–200bp);
[0071] 4) Try to avoid primer-dimer and hairpin structures;
[0072] 5) Too high (>70%) or too low (<30%) GC content is not conducive to RPA amplification;
[0073] 6) For the three nucleotides at the 3' end, guanine and cytosine contribute to the stable combination of the polymerase, which can improve the amplification perform...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com