Soybean identity important character SNP (single-nucleotide polymorphism) marker combined screening method and application thereof
A technology of soybeans and soybeans, which is applied in the direction of biochemical equipment and methods, and the measurement/inspection of microorganisms, can solve the problems of difficult existing varieties, accurate identification, etc., and achieve the effect of wide identification and adaptability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1、14
[0041] Example 1, Screening of 14 SNP site combinations GlySNP14
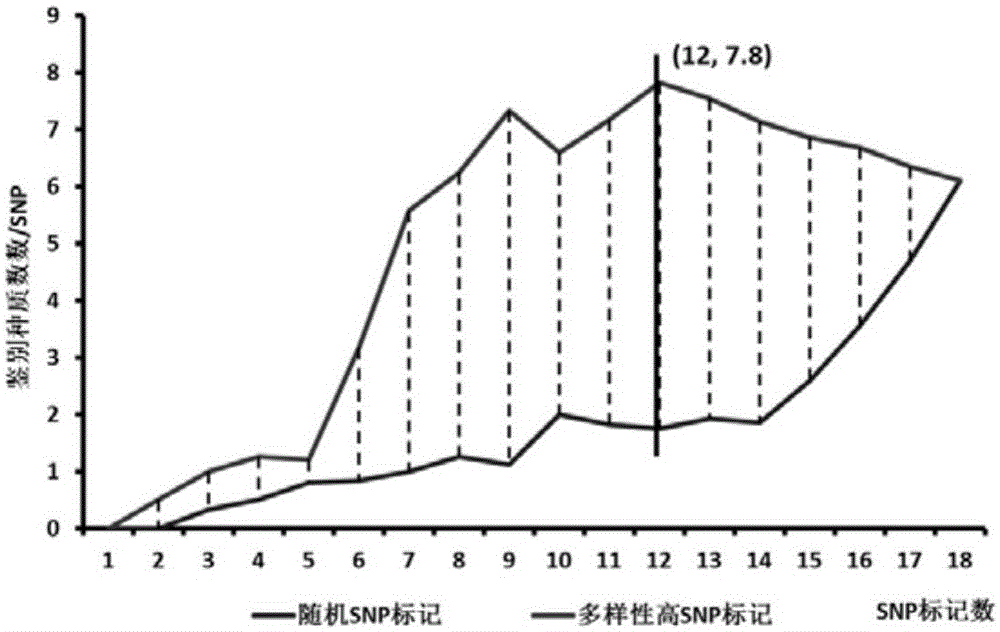
[0042] 254 phenotype-accurately identified germplasms from Northeast China and Huanghuai were used for SNP marker identification of important traits, including 102 northern germplasms and 152 Huanghuai germplasms (Table 1). By optimizing different combinations of SNP markers, the soybean germplasm ID card was finally constructed with the least number of SNP markers.
[0043] Table 1 shows the SNP sites of 254 soybeans
[0044]
[0045]
[0046]
[0047]
[0048]
[0049]
[0050]
[0051]
[0052]
[0053]
[0054]
[0055]
[0056]
[0057]
[0058]
[0059] 1. Screening of 14 SNP sites
[0060] 1) 23 SNP markers
[0061] In the present invention, 23 SNP markers come from 12 genes altogether, which are respectively related to pod-setting habit, hair color, ripening period, hundred-grain weight, soybean cyst nematode disease and mosaic virus disease resistance (Tab...
Embodiment 2、14
[0082] Example 2, 14 SNP loci combined with GlySNP14 to identify 60 northern soybean germplasms
[0083] 1. Extract soybean genomic DNA
[0084] Genomic DNA of 60 northern soybean leaves in Table 3 was extracted.
[0085] 2. Detection
[0086] The bases of the 14 SNP sites in the above soybean genomic DNA were detected by the mass spectrometry detection SNP typing (instrument: equinomMassarray genotyping system) technology of Bioyong Technologies Inc.
[0087] As a result, the bases of each SNP site of each soybean are shown in Table 3.
[0088] Table 3 is the nucleotides of 14 SNP sites on 60 northern soybean leaves and genome
[0089]
[0090]
[0091]
[0092]
[0093]
[0094] Using GlySNP14 to identify 60 northern soybean germplasms, the genetic diversity index of each SNP marker varied from 0 to 0.939, with an average value of 0.523. Among the 14 SNP markers, 7 SNPs had transversions, and the variation range was 0.451-0.918; 2 SNPs were deleted, and the ...
Embodiment 3、14
[0099] Example 3. Combination of 14 SNP loci GlySNP14 to identify 36 Huanghuaihai soybean germplasms
[0100] 1. Extract soybean genomic DNA
[0101] Genomic DNA of 36 Huanghuaihai soybean leaves in Table 4 was extracted.
[0102] 2. Detection
[0103] The bases of the 14 SNP sites in the above soybean genomic DNA were detected by the mass spectrometry detection SNP typing (instrument: equinomMassarray genotyping system) technology of Bioyong Technologies Inc.
[0104] As a result, the bases of each SNP site of each soybean are shown in Table 4.
[0105] Table 4 shows the nucleotides of 14 SNP sites on 36 Huanghuaihai soybean leaves and genome
[0106]
[0107]
[0108]
[0109]
[0110] Using GlySNP14 to identify 36 Huanghuaihai soybean germplasms, the genetic diversity index of each SNP marker varied from 0 to 0.693, with an average value of 0.485. Among the 14 SNP markers, 8 SNPs had transversions, and the variation range was 0.349-0.668; 2 SNPs were deleted...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com