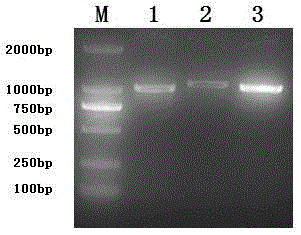
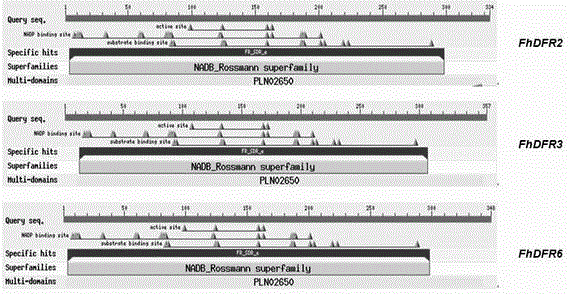
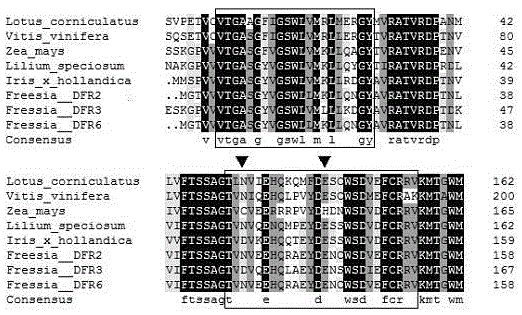
CDNA of Freesia refracta Klatt flavanonol-4-reductase genes
A technology of vanilla dihydroflavonol and reductase, which is applied in the field of cDNA and can solve problems such as functions that have not been verified
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Example 1: FhDFRs Cloning of full-length genes
Embodiment 1-1
[0039] Example 1-1: FhDFRs Acquisition of 3' end: According to the intermediate fragment obtained by screening, design primers for 3' RACE-PCR amplification. The primer combinations are:
[0040]
[0041] The reaction system is as follows:
[0042]
[0043] The PCR reaction procedure is as follows: 95°C for 5min; 94°C for 30s, X°C for 30s, 72°C for 1min, 30Cycle.
[0044] where X°C is the annealing temperature, FhDFR2 (58°C) ,FhDFR3 (58°C) , FhDFR6 (58°C).
[0045] After the gel recovery of the PCR product, it was connected to the T vector and transformed into Escherichia coli JM109Competent cells were screened with Amp-resistant LB plate medium. Randomly pick a single clone, shake the bacteria and carry out the PCR verification of the bacterial liquid, and the positive clones identified are sequenced.
Embodiment 1-2
[0046] Embodiment 1-2: FhDFRs Acquisition of the 3' end: splicing and screening the intermediate fragment and the amplified 3' end fragment, and then designing primers based on this, and using the RACE kit (SMARTRACE cDNA Amplification Kit) from Clontech to perform 5' RACE experiments.
[0047] Primer combinations are as follows:
[0048]
[0049] The reaction system is as follows:
[0050] ① FhDFR2 The touchdown PCR reaction
[0051] The reaction system is as follows:
[0052]
[0053] The reflection procedure is as follows:
[0054]
[0055] ② FhDFR3 nested PCR reaction
[0056] A. Using DFR3-GSP2 as the downstream primer for PCR, the reaction system is as follows:
[0057]
[0058] The reaction procedure is as follows:
[0059]
[0060] B. Dilute the above PCR product 50 times as a template, and use DFR3-GSP1 and NUP as nested primers for PCR
[0061] Amplify. The reaction system system is as follows:
[0062]
[0063] The reaction procedure ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com