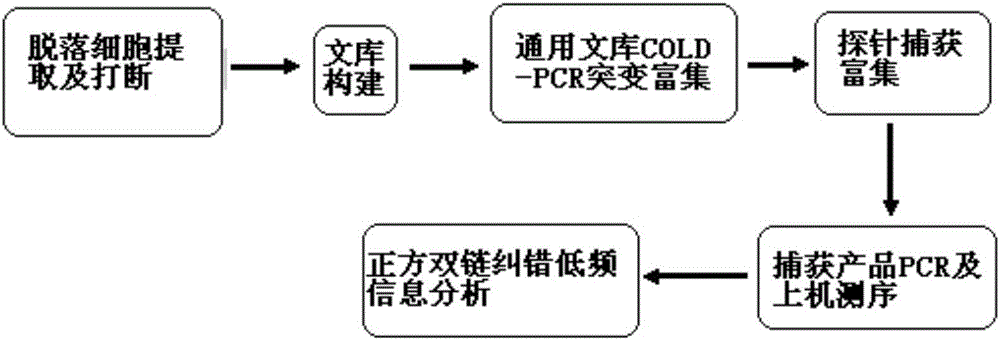
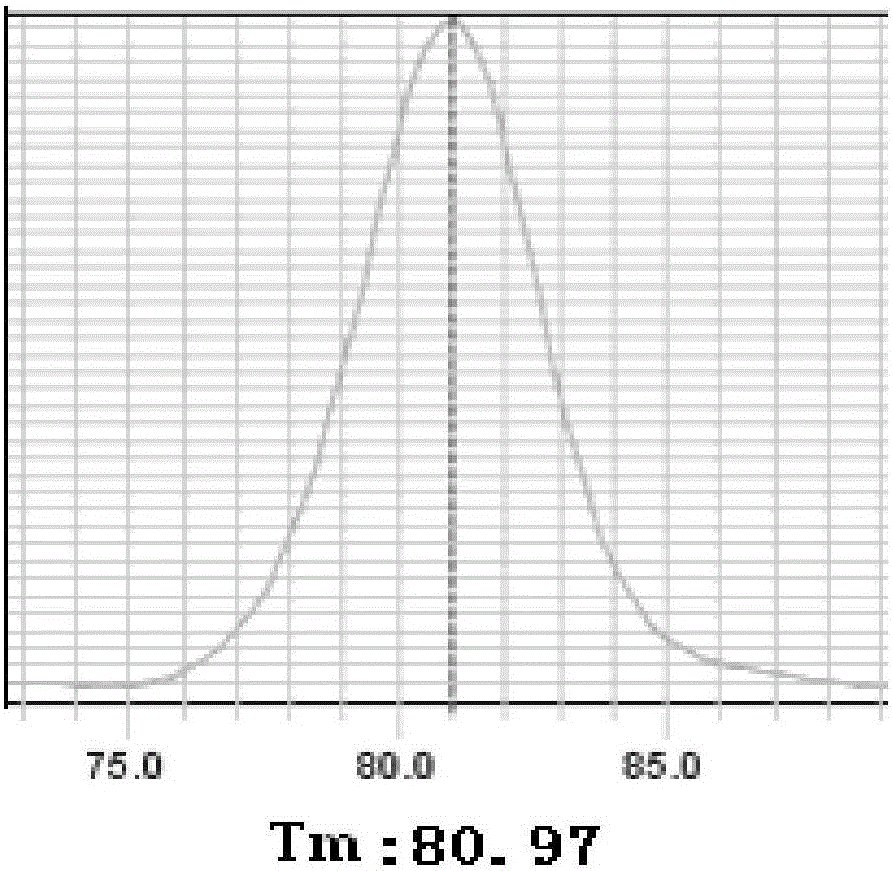
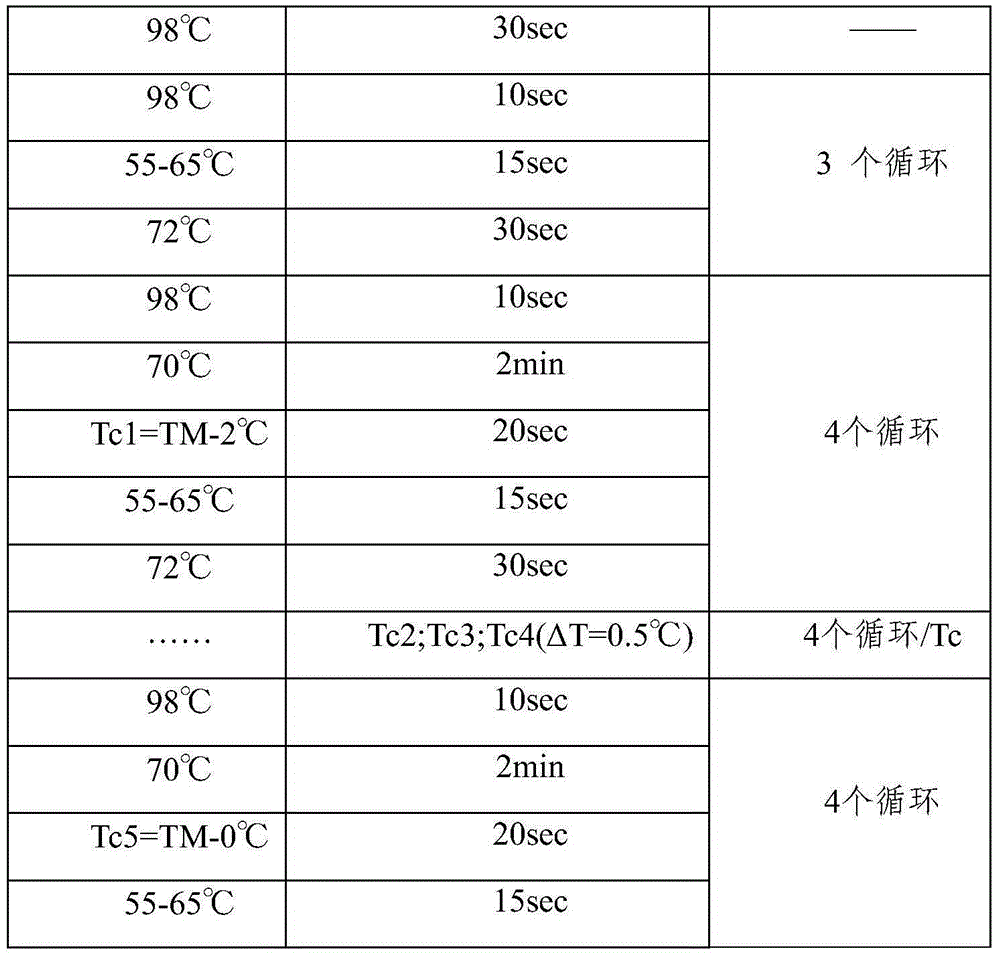
Method for low-frequency mutant-enriched sequencing of DNA of exfoliative cells
A technique of shedding cells and low-frequency mutations, applied in the field of high-throughput sequencing in bioinformatics, which can solve problems such as laborious, fluctuating, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0096] Example 1 Establishment of DNA Low-Frequency Mutation Enrichment Sequencing Method for Exfoliated Cervical Cells
[0097] 1. Extraction and interruption of DNA from cervical exfoliated cells:
[0098] Aspirate 1-2ml of the patient's cervical exfoliated cell mixture, centrifuge at 16000g for 10 minutes at 4°C, and remove the supernatant. The isolated precipitated cells were extracted from exfoliated cells based on the instructions of the QIAampDNAMiniKit (QIAGEN) extraction reagent, and the DNA was extracted according to the kit Qubit (Invitrogen, theQuant-iTTMdsDNABRAssayKit). Take 1 μg of DNA and cut into 200-400 bp fragments according to the 100 μL disruption system based on the instruction manual of the BioruptorPico disruption instrument. The disruption conditions are: 10min / H, ON=30s; OFF=30s; cycle=10.
[0099] 2. Preparation of the sample library: After the fragmented exfoliated cell DNA, a 3-step enzymatic reaction was carried out according to the KAPALTPLibrar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com