Method for detecting beta-casein genotype on basis of restriction enzyme digestion
A detection method, casein technology, applied in the direction of biochemical equipment and methods, microbial determination/inspection, etc., can solve the problems of unguaranteed and inability to detect different genotypes of β-casein
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] Use primers to carry out PCR amplification of the dairy cow genomic DNA to be detected: take 1ul of DNA template (45ng / ul), add 0.5ul of 0.1U / ul hot-start TaqDNA polymerase, 2.5uM of dNTP1ul, 0.5 of each of 10uM upstream and downstream primers ul, 2×PCRBuffer 8ul, add double distilled water to 20ul to obtain a PCR amplification system, heat the amplification system to 95°C for 5 minutes, then denature at 95°C for 15s, anneal at 52°C for 35s, extend at 68°C for 35s, and self-denature One cycle was counted from the beginning to the end of the extension, and after 30 cycles, the extension was performed at 68° C. for 10 min.
[0050] Use the restriction endonuclease TaqI to digest the PCR amplification product: Take 5ul of the PCR amplification product, add restriction endonuclease TaqI0.05ul, buffer 0.5ul, incubate at 60°C for 4h, heat inactivate at 75°C 30min.
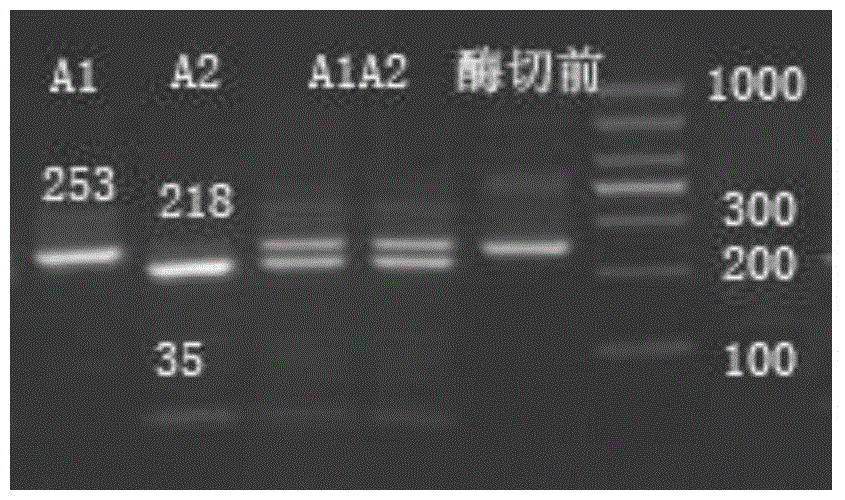
[0051] Test results such as figure 1 As shown, the amplified product will produce a 218bp fragment and a 35bp...
Embodiment 2
[0053] Utilize primers to carry out PCR amplification of the dairy cow genomic DNA to be detected, take DNA template 1ul (50ng / ul), add 0.1U / ul hot-start TaqDNA polymerase 1ul, 2.5uM dNTP1ul, 10uM upstream and downstream primers each 1ul, 2×PCRBuffer10ul, add double-distilled water to 20ul to obtain a PCR amplification system, heat the amplification system to 95°C for 5 minutes, then denature at 95°C for 15s, anneal at 61°C for 30s, and extend at 72°C for 30s. The end of the extension was counted as one cycle, and after 35 cycles, it was extended at 72° C. for 7 min.
[0054] Use restriction endonuclease TaqI to carry out enzyme digestion, judge according to the result of agarose gel electrophoresis, take 9ul of PCR amplification product, add restriction endonuclease TaqI0.2ul, buffer solution 1ul, incubate at 65 ℃ for 3h, in Heat inactivation at 80°C for 20min.
[0055] Test results such as figure 1 As shown, the amplified product will produce a 218bp fragment and a 35bp sm...
Embodiment 3
[0057] Use primers to perform PCR amplification of the cow genomic DNA to be detected, take 1ul of DNA template (55ng / ul), add 0.1U / ul hot start TaqDNA polymerase 1.5ul, 3uM dNTP1.5ul, 15uM / ul upstream and downstream 1.5ul of each primer, 12ul of 2×PCRBuffer, add double distilled water to 20ul to obtain a PCR amplification system, heat the amplification system to 95°C for 5min, then denature at 92-95°C for 15s, anneal at 65°C for 25s, and 72°C Extend for 25 s, and count as one cycle from the beginning of denaturation to the end of extension, and after 45 cycles, extend at 72°C for 4 min.
[0058] Use restriction endonuclease TaqI to carry out enzyme digestion, judge according to the result of agarose gel electrophoresis, take PCR amplification product 13ul, add restriction endonuclease TaqI0.35ul, buffer 1.5ul, incubate at 65 ℃ for 3h, Heat inactivation at 80°C for 20min.
[0059] Test results such as figure 1 As shown, the DNA amplification product containing A1β-casein cow...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com