Kit for detecting HBV through bulk sample PCR, and application thereof
A technology of kits and reagents, applied in the field of kits for detection of HBV by sample collection PCR, can solve the problems of repeatability and stability, and the cost of kits does not have advantages, so as to ensure the detection rate, reduce sample processing time and Steps, the effect of saving reagent expenses
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0028] Embodiment 1, the design of primer
[0029] According to the comparison of HBV conserved sequences, two pairs of degenerate primers S1-F / S1-R and S2-F / S2-R were designed to establish a nested PCR method for HBV detection. The specific sequences are listed in Table 1;
[0030] Table 1 is the HBVPCR amplification primer sequence
[0031]
Embodiment 2
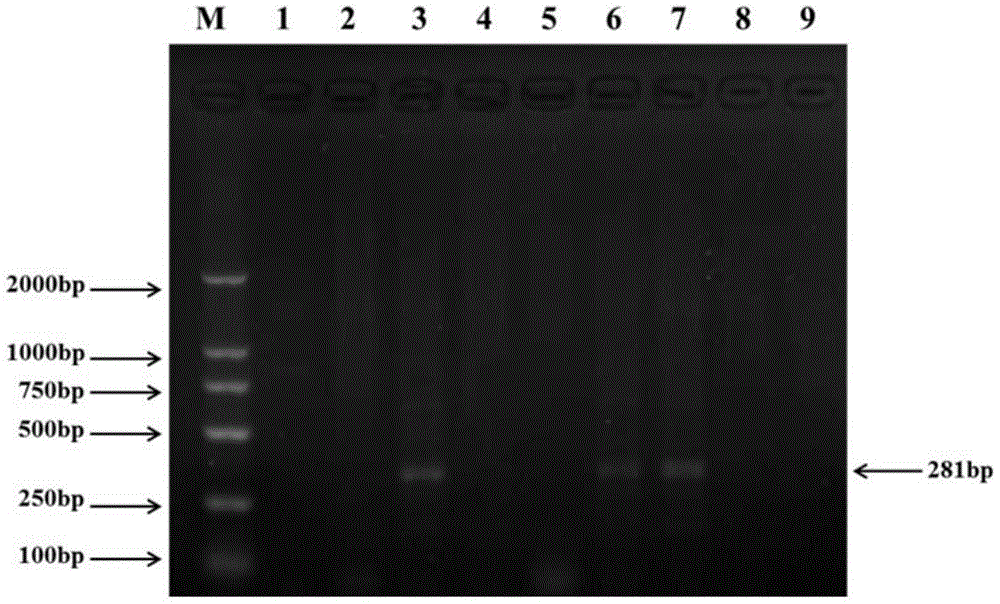
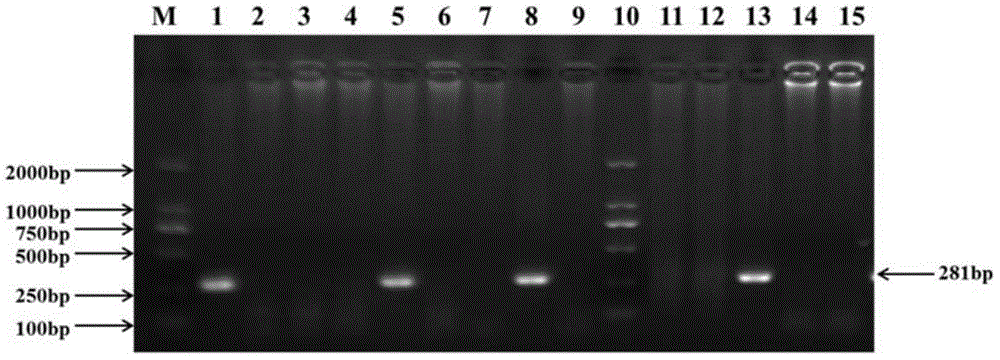
[0032] Embodiment 2, primers are used for PCR detection of HBV by collection sample method
[0033] 1. Preparation of sample to be tested by collection method
[0034] A total of 32 liver samples from slaughtered chickens were collected, marked and stored at -80°C.
[0035] Put about 100 mg of each of the 4 samples to be tested in a mortar, add liquid nitrogen to grind and mix them evenly, and then use a 1.5ml centrifuge tube to collect samples and mix and grind about 100 mg of tissue for follow-up tests. There are 8 groups of mixed samples to be tested. The mixed samples of each sample set shall be marked for secondary detection.
[0036] One group: numbered 13-16, two groups: numbered 17-20, three groups: numbered 1-4, four groups: numbered 21-24, five groups: numbered 25-28, six groups: numbered 5-8, seven groups: numbered 9-12, eight groups: numbered 29-32. Among the above 32 samples, the samples coded as 1, 5, 8, and 13 were known to be infected with HBV, and the rest ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com