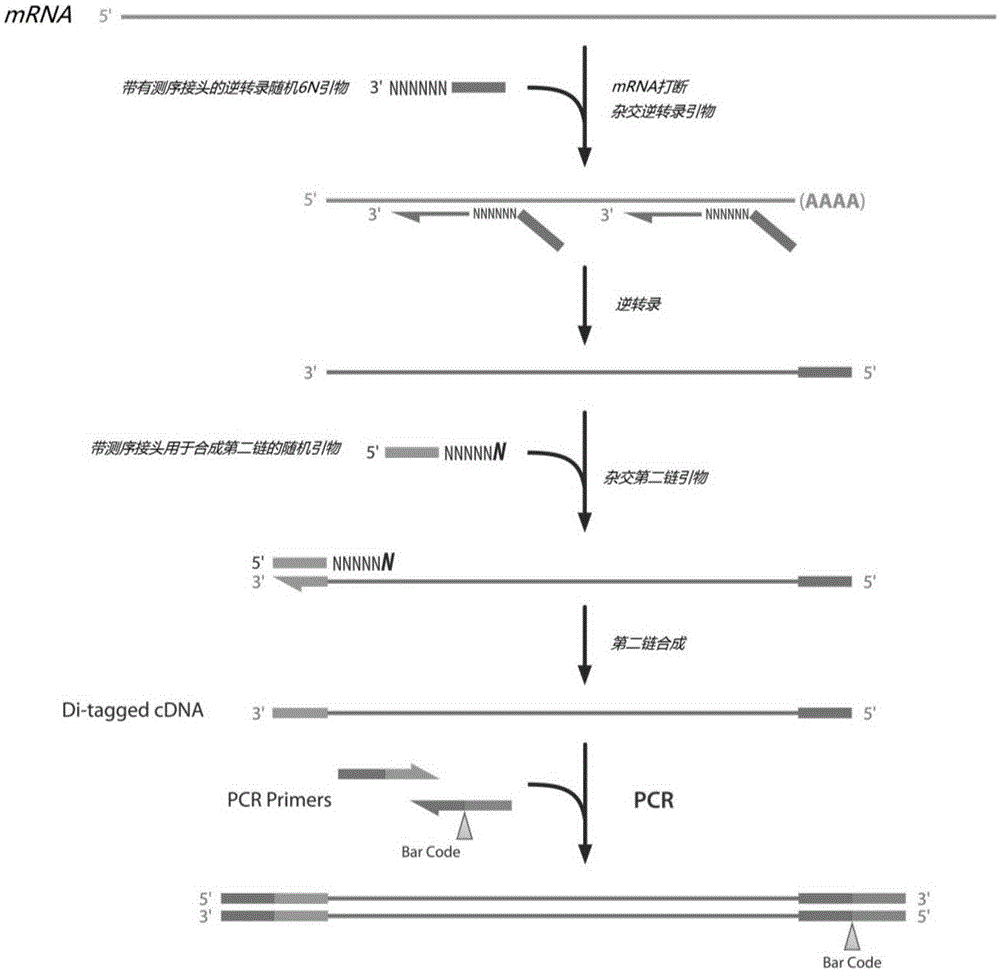
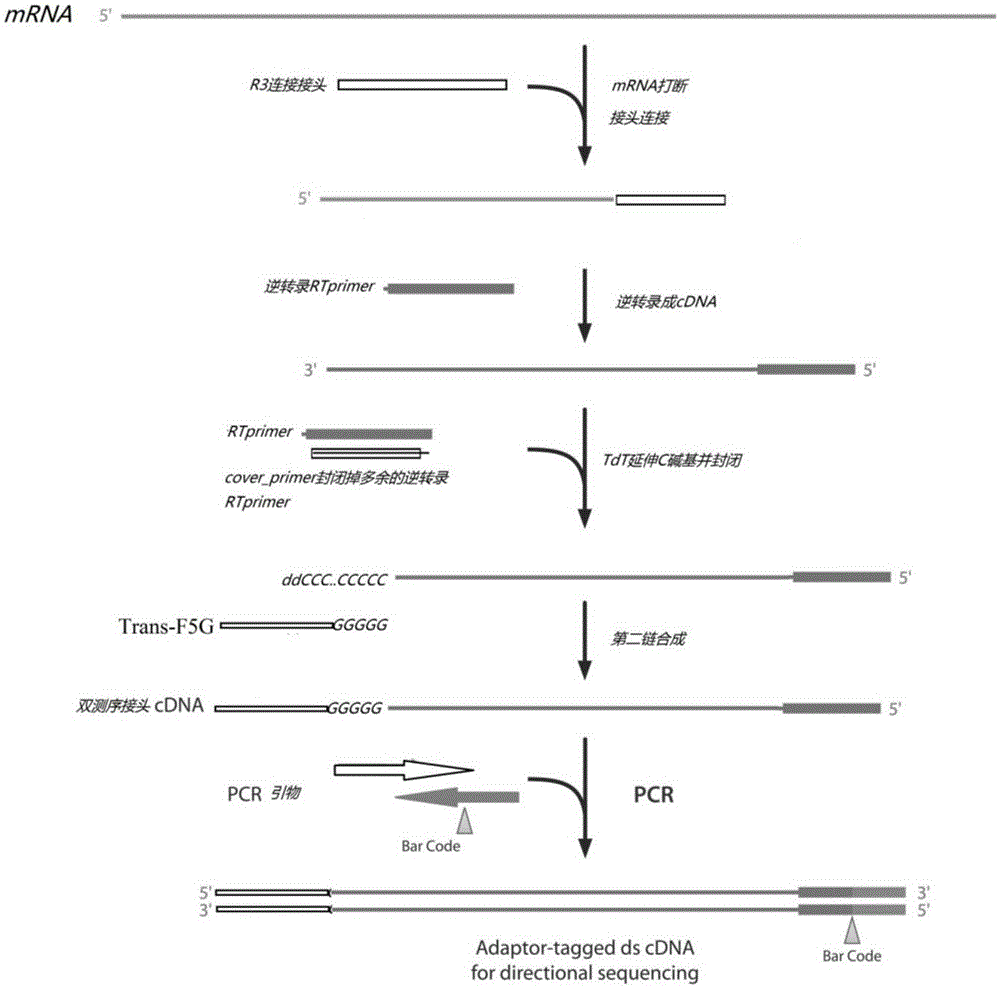
Method for constructing strand-specific transcriptome library
A transcriptome-specific technology, applied in the field of constructing strand-specific transcriptome libraries, can solve the problems of low labeling efficiency at the 3' end of cDNA, facilitate data analysis, improve cDNA synthesis efficiency, and reduce the production of linker dimers. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] mRNA fragmentation treatment: Qualified RNA was extracted, and mRNA was purified by mixing 2550 μL of Dynabeadsoligo (dT) from life-tech company with 1 μg of total RNA. Take 7 μL of 5× (that is, 5 times) fragmentbuffer and add it to 28 μL of mRNA, 94°C for 5 minutes, and then put it on ice immediately. Add 65 μLddH 2 O (i.e. double distilled water), 20 μg glycogen, 1 / 10 volume of 3M pH5.2 NaAc, 2.5 times volume of absolute ethanol, mix well, store at -80°C for 20 min; centrifuge at high speed, and dissolve the precipitate in 13 μL deionized water.
[0028] Ligation of R3 linker: mRNA dissolved in 5 μL ddH2 In O, add 1 μL of 10 μM R3 linker, 70 ° C for 2 min, and place on ice for 1 min. Add 1.5μL ligationMix, 200UT4 truncated RNA ligase 2K227Q (T4RNAligase2TruncatedK227Q), ddH 2 Make up to 15 μL with O, mix well, and keep at 22°C for 1h.
[0029] cDNA synthesis: Add 1 μL of 50 μM RTprimer, 65°C for 5 min, and place on ice for 1 min; add 4 μL of RTMix, 20 UMMLV reverse...
Embodiment 2
[0034] mRNA fragmentation treatment: Qualified RNA was extracted, and mRNA was purified by mixing 2530 μL of Dynabeadsoligo (dT) from life-tech company with 1 μg of total RNA. Take 7 μL of 5×fragmentbuffer and add it to 28 μL of mRNA, 94°C for 5 minutes, and then put it on ice immediately. Add 65 μLddH 2 O, 20 μg glycogen, 1 / 10 volume of 3M NaAc at pH 5.2, 2.5 volumes of absolute ethanol, mix well, store at -80°C for 20 min; centrifuge at high speed, and dissolve the precipitate in 13 μL deionized water.
[0035] Ligation of R3 linker: mRNA dissolved in 5 μL ddH 2 In O, add 0.5 μL of 10 μM R3 adapter, 70 ° C for 2 min, and place on ice for 1 min. Add 1.5 μL of ligationMix, 200UT4RNAligase2TruncatedK227Q, ddH 2 Make up to 15 μL with O, mix well, and keep at 22°C for 1h.
[0036] cDNA synthesis: Add 0.5 μL of 50 μM RTprimer, 65° C. for 5 minutes, and place on ice for 1 minute. Add 4 μL RTMix, 20 UMMLV reverse transcriptase, ddH 2 Make up to 20ul with O, mix well, then stor...
Embodiment 3
[0046] mRNA fragmentation treatment: Qualified RNA was extracted, and mRNA was purified by mixing 2550 μL of Dynabeadsoligo (dT) from life-tech company with 1 μg of total RNA. Take 7 μL of 5Xfragmentbuffer and add to 28 μL of mRNA, 94°C for 5 minutes, and then put it on ice immediately. Add 65 μLddH 2 O, 20 μg glycogen, 1 / 10 volume of 3M pH5.2NaAc, 2.5 times the volume of absolute ethanol, mix well, store at -80°C for 20min; centrifuge at high speed, and dissolve the precipitate in 13 μL deionized water.
[0047] Ligation of R3 linker: mRNA dissolved in 5 μL ddH 2 In O, add 1 μL of 10 μM R3 linker, 70 ° C for 2 min, and place on ice for 1 min. Add 1.5 μL of ligationMix, 200UT4RNAligase2TruncatedK227Q, ddH 2 Make up to 15 μL with O, mix well, and keep at 22°C for 1h.
[0048] cDNA synthesis: Add 1 μL of 50 μM RTprimer, 65°C for 5 min, and place on ice for 1 min; add 4 μL of RTMix, 20 UMMLV reverse transcriptase, ddH 2 Make up to 20 μL with O, mix well, then store at 25°C f...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com