Method for detecting microRNA (microribonucleic acid) on basis of rolling circle amplification and upconversion material
A technology for converting materials and detecting probes, applied in the fields of molecular biology and nucleic acid chemistry, can solve the problem of high DNA dye background, and achieve the effects of overcoming light scattering, good sequence selectivity, and long fidelity time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
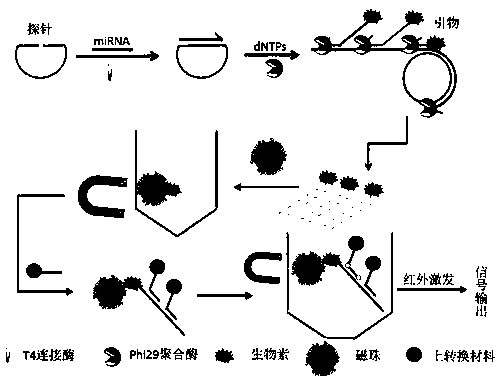
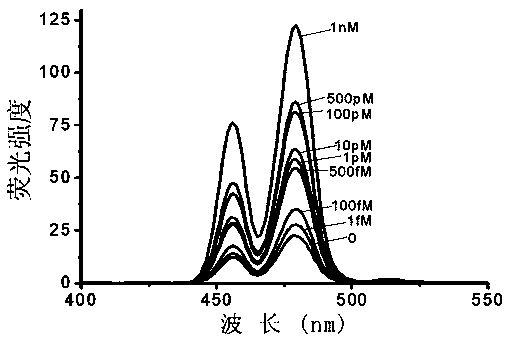
[0028] 1. Design of microRNAs detection system
[0029] (1) if figure 1 As shown, the microRNAs detection probes used in the examples were synthesized by Shanghai Sangong Co., Ltd., and the probes were synthesized against the microRNAslet-7a sequence. The sequence of the microRNAs is: UGAGGUAGUAGGUUGUAUAGUU; the 3' end segment of the microRNA detection probe has 11 bases are complementary paired with the 5' end of the target microRNA, 11 bases are complementary paired with the 3' end of the target microRNA in the 5' end segment, and the middle segment is single-stranded DNA. The probe sequence from 5' end to 3' end is: phospho-CTACTACCTCATTTGCATTTCAGTTTACGGTTTAGCATTTCGCAATTTTAACTATACAAC. The added primer is a DNA sequence modified with biotin at the 5' end, which is exactly the same as a sequence in the probe. The primer sequence is: biotin-CAACCTACTACCTCATTTGC. In addition, the DNA modified on the upconversion material is characterized by the DNA sequence modified by the am...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com