Amplification method for whole genome aiming at different enterovirus serotypes
An enterovirus and whole genome technology is applied in the field of amplification of whole genomes of different enterovirus serotypes. The amplification method is simple and easy, the rapid amplification method, and the effect of fast amplification speed
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Taking Coxsackievirus B1 (CV-B1) as an example
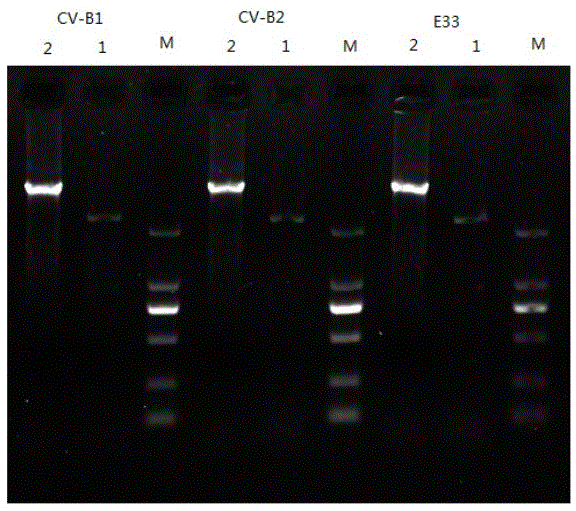
[0024] (1) Perform RT-PCR on the VP1 region of the viral RNA with enterovirus general primer 222 (5'CICCIGGIGGIAYRWACAT3')-224 (GCIATGYTIGGIACICAYRT), and then use 1% agarose gel for electrophoresis detection at a voltage of 120V, and the detection results Compared with the predicted results, the results are similar (see figure 2 ), after sequencing, use NCBIBLAST (a set of analysis tools for similarity comparison in the protein database or DNA database of the National Center for Biotechnology Information of the United States, and the BLAST program can quickly compare similarity sequences with public databases) to compare, the nucleoside The homology between the acid sequence and Coxsackie virus B1 is 92%. According to the typing standard, it can be judged that the virus is Coxsackie virus B1 (CV-B1). This amplification obtained part of the VP1 in the genome of the virus. the nucleotide sequence of the gene; ...
Embodiment 2
[0042] Example 2: Taking Coxsackievirus B2 (CV-B2) as an example
[0043] (1) Perform RT-PCR on the VP1 region of the viral RNA with enterovirus general primers 222-224, and then use 1% agarose gel for electrophoresis detection at a voltage of 120V. The detection results are compared with the predicted results, and the results are similar ( See figure 2 ), compared with NCBIBLAST after sequencing, the nucleotide sequence has a homology of 92% with CV-B2. According to the typing standard, the virus can be judged as CV-B2, and part of the VP1 in the genome of the virus can be obtained The nucleotide sequence of the gene.
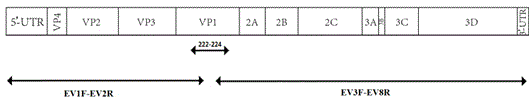
[0044] (2) According to the conserved type of the 5' end and 3' end of the enterovirus genome, and the nucleotide sequence of the part of the VP1 gene obtained in step (1), design primers; divided into two sections, the designed primers are: EV1F-B22R ( About 2400bp), B23F-EV8R (about 5000bp) (see Table 4), using viral RNA as a template, EV1F-B22R (about 24...
Embodiment 3
[0061] Example 3: Taking echovirus type 33 (E-33) as an example
[0062] (1) Perform RT-PCR on the VP1 region of the viral RNA with enterovirus general primers 222-224, and then use 1% agarose gel for electrophoresis detection at a voltage of 120V. The detection results are compared with the predicted results, and the results are similar ( See figure 2 ), compared with NCBIBLAST after sequencing, the nucleotide sequence has a homology of 83% with E-33. According to the typing standard, it can be judged that the virus is E-33, and part of the VP1 in the genome of the virus can be obtained the nucleotide sequence of the gene;
[0063] (2) According to the conserved type of the 5' end and 3' end of the enterovirus genome, and the nucleotide sequence of part of the VP1 gene obtained in step (1), design primers; in two sections, design primers: EV1F–E332R (approx. 2400bp), E333F-EV8R (about 5000bp) (see Table 6), using viral RNA as a template, EV1F-E332R (about 2400bp), E333F-EV...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com