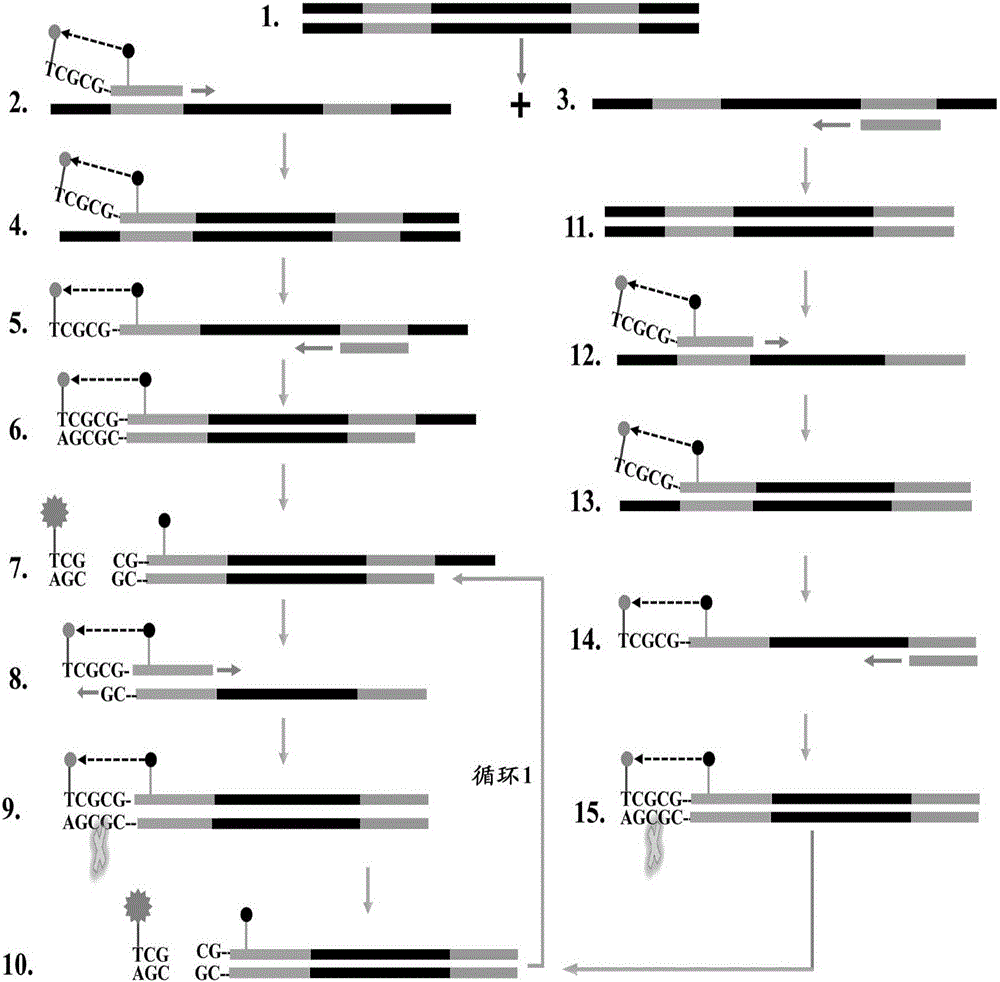
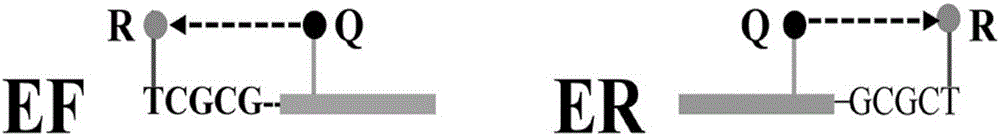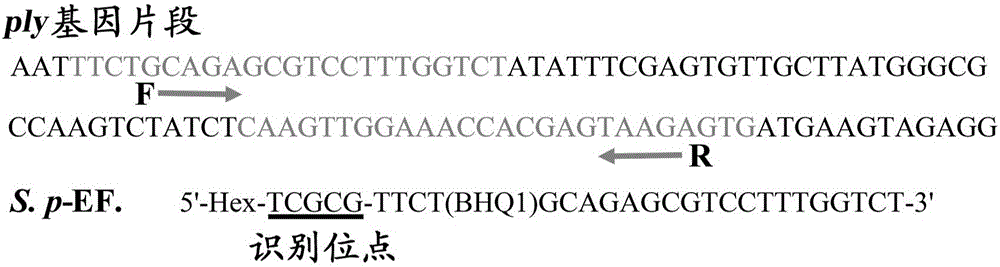ET-PCR (endonuclease restriction-mediated real-time polymerase chain reaction) nucleic acid testing technology
A technology of restriction endonuclease and amplification system, which is applied in the determination/inspection of microorganisms, methods based on microorganisms, biochemical equipment and methods, etc., and can solve the problems of poor repeatability of results, easy misjudgment, time-consuming and labor-intensive, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0059] Embodiment 1 standard ET-PCR reaction
[0060] 1. Standard ET-PCR reaction system:
[0061]
[0062] 2. Standard ET-PCR reaction amplification parameters
[0063] Amplify and measure the results on a real-time fluorescent quantitative PCR instrument:
[0064]
[0065] The Real-time reaction was carried out in a fluorescence detector (Rotor-Gene Q Real Time System, Qiagen). After the amplification was completed, the background fluorescence signal was subtracted and the data was analyzed at the same threshold to determine the Ct (cyclethreshold) value of the reaction.
[0066] 3. Feasibility of ET-PCR amplification reaction
[0067] (1) Fluorescence detection method: Under standard ET-PCR reaction conditions, ET-PCR generates a large number of fluorescent signals when amplifying DNA, which can be detected by a real-time fluorescence detector and generate stable fluorescent signals. The intensity correlates positively with the product of the amplicon, see Figure...
Embodiment 2
[0078] Embodiment 2: multiplex ET-PCR reaction
[0079] 1. Multiplex ET-PCR reaction system:
[0080]
[0081] 2. Multiplex ET-PCR amplification parameters
[0082] Amplify and measure the results on a real-time fluorescent quantitative PCR instrument:
[0083]
[0084]
[0085] The Real-time reaction was carried out in a fluorescence detector (Rotor-Gene Q Real Time System, Qiagen). After the amplification was completed, the background fluorescence signal was subtracted and the data was analyzed at the same threshold to determine the Ct (cyclethreshold) value of the reaction.
[0086] 3. The multiple detection ability of ET-PCR
[0087] In order to obtain stable multiple fluorescence detection patterns, the standard ET-PCR system was optimized to adapt to multiple detection, and a multiple detection system was established. Under the multiplex detection system, we added three sets of primers and corresponding templates to the multiplex reaction mixture at the same ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com