A high-efficiency plasmid system for allele integration of Saccharomyces cerevisiae and its application
An allele, Saccharomyces cerevisiae technology, applied in the direction of using vectors to introduce foreign genetic material, microorganisms, nucleic acid vectors, etc., to achieve a wide range of applications, high-throughput allele integration, and improvement of yeast gene function related research technology effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
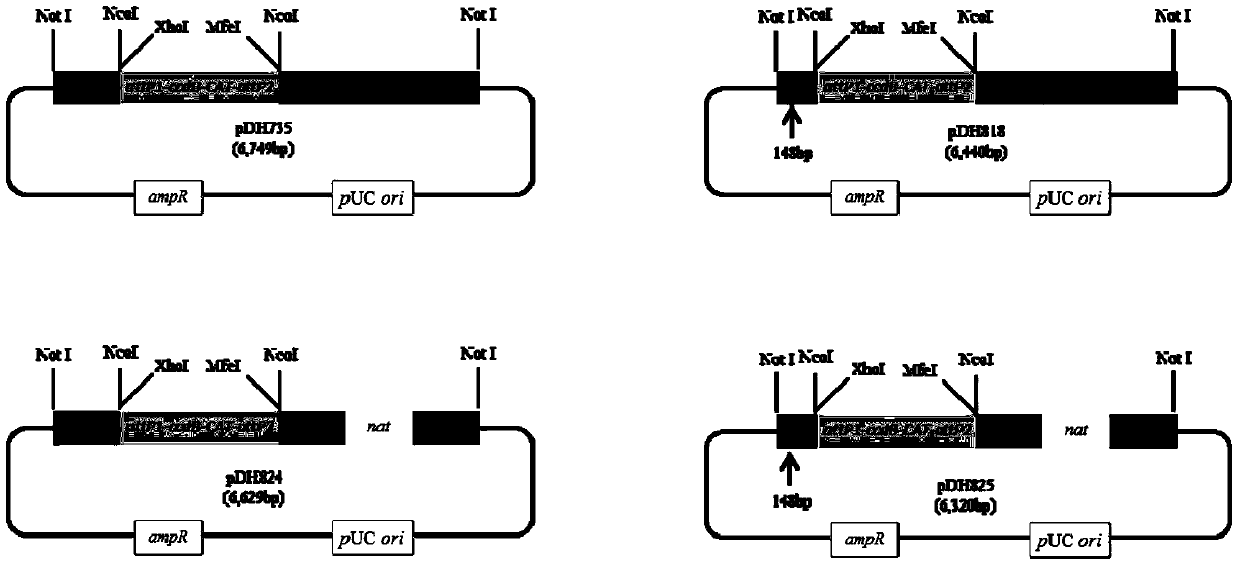
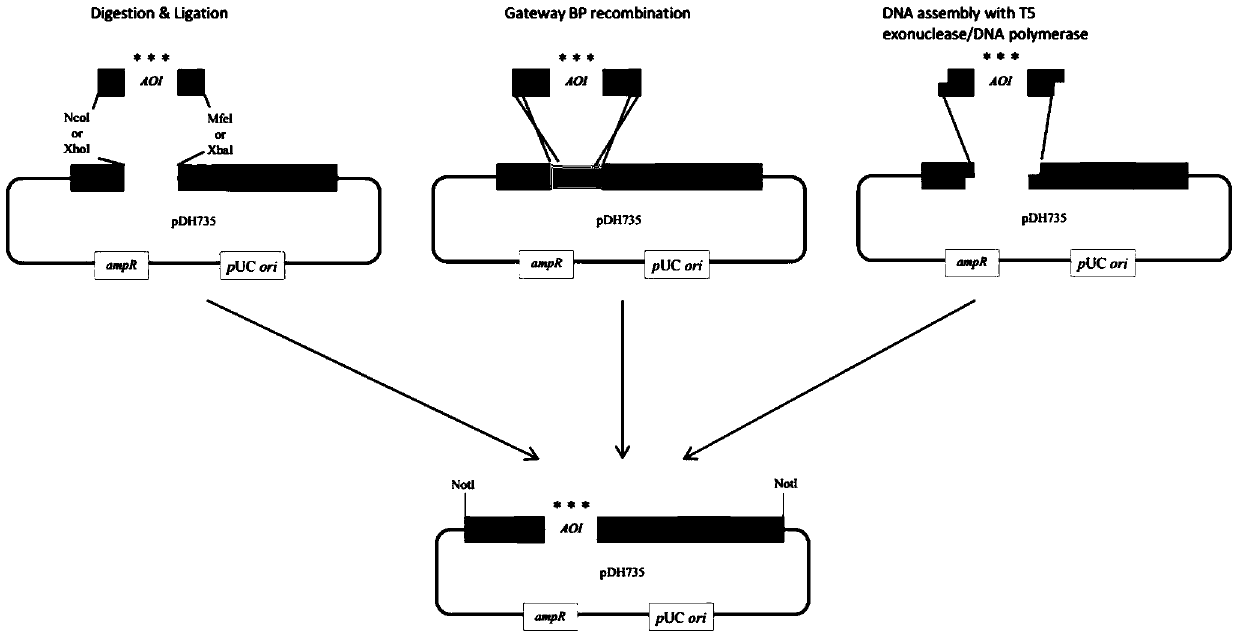
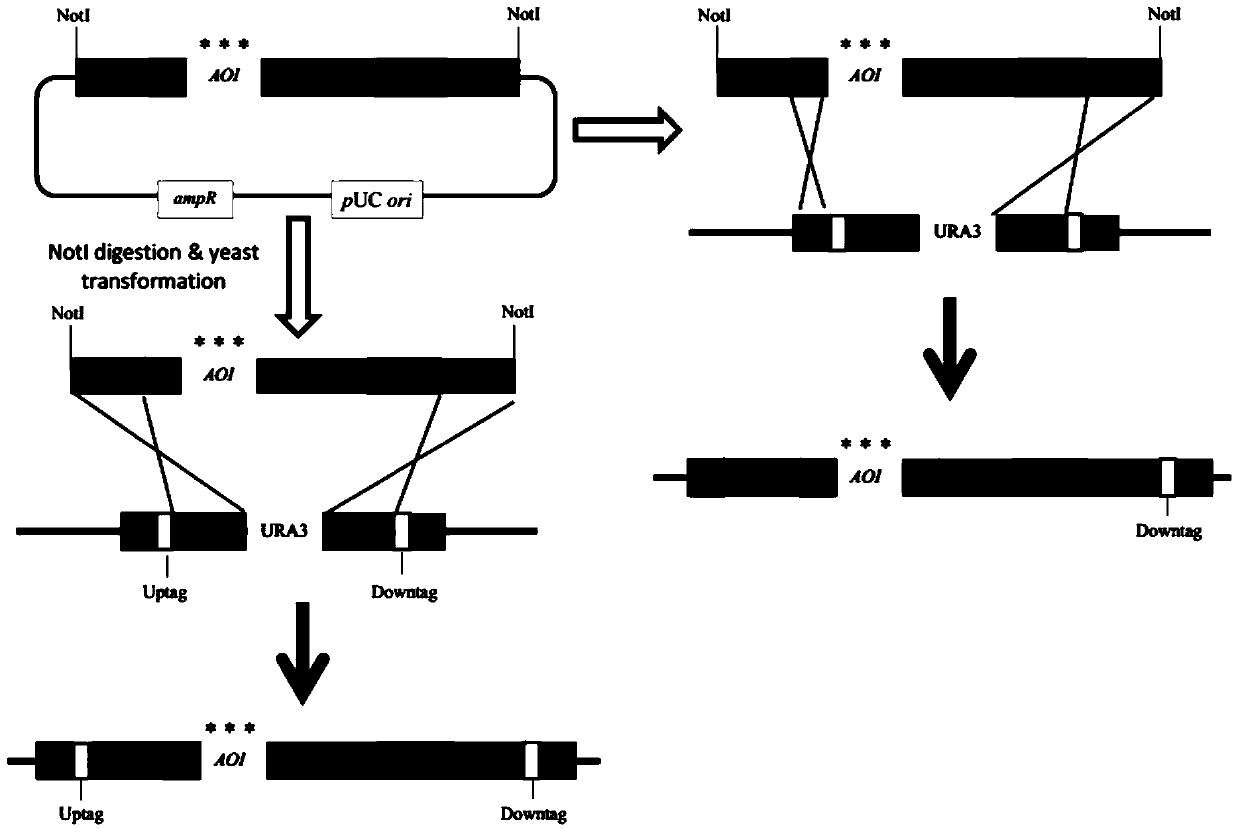
[0034] Construction of plasmid pDH735.
[0035] 1. Design primers
[0036] EM7-F: TCTCACATCACATCCGAACATAAACAACCATGGTCAGTCTAGATGTTGACAATTAATCATCGGC
[0037] EM7-R: TGCACTTCTCAGTACAATCTGGTTTAG TTCCTCACCTTGTC
[0038] TRP1-F:GACAAGGTGAGGAACTAAACC AGATTGTACTGAGAGTGCA
[0039] TRP1-R: CCTCGAAACGTGAGTCTTTTTCCTTACCCATACTCCAAGCTGCCTTTGTGT
[0040] ATTP1-F: CATGCCATGGCTCGAGCAAATAATGATTTTATTTTGACTGA
[0041] ATTP1-R:GCGTGGATCCGGCTTACTA
[0042] ATTP2-F:TAGTAAGCCGGATCCACGC
[0043] ATTP2-R:GCTCTAGACAATTGCAAATAATGATTTTATTTTGACTGA
[0044] 2. Using yeast genomic DNA as a template and EM7-F and EM7-R as amplification primers, use a conventional PCR system and reaction program for PCR amplification to obtain the PCR product of the EM7 promoter. Similarly, the PCR product of the TRP1 promoter was amplified by using yeast genomic DNA as a template and TRP1-F and TRP1-R as amplification primers. Then the PCR products of EM7 promoter and TRP1 promoter were purified and recovered.
[004...
Embodiment 2
[0049] Construction of plasmid pDH818.
[0050] 1. Design primers
[0051] PT-F:GAATTCTCGAGCCTAGGACTGTCAAGGAGGGTATTC
[0052] PT-R:GAATTCTCGAGTAAGGAAAAGACTCACGTTTC
[0053] 2. Using the plasmid pFA6a-kanMX as a template, PT-F and PT-R as amplification primers, use conventional PCR system and reaction procedures to perform PCR amplification to obtain the PCR product of pFA6a-kanMX, and then purify and recover the PCR product.
[0054] 3. The purified PCR product of pFA6a-kanMX is digested with XhoI, and the digested product is further purified and recovered.
[0055] 4. The purified digested product was self-ligated under the action of T4 ligase, and the correct plasmid was obtained after screening with kanamycin, and the obtained plasmid was named pDH817.
[0056] 5. Carry out XhoI / SacI double digestion of plasmids pDH735 and pDH817 to obtain the attP1-ccdB-CAT-attP2-pTRP1 cassette and linearized pDH817. The digested products are purified and catalyzed by T4 ligase, and the...
Embodiment 3
[0058] Construction of plasmid pDH824.
[0059] 1. Design primers
[0060] ADH1-F:
[0061] TTCTCACATCACATCCGAACATAAACAACCATGGTCAGTCTAGACTTGATAGCCATCATCATATC
[0062] ADH1-R: GTAAGCCGTGTCGTCAAGAGTGGTACCCATTGTATATGAGATAGTTGATTG
[0063] 2. Using yeast genomic DNA as a template, ADH1-F and ADH1-R as amplification primers, clone the ADH1 promoter into pAG25 according to the method in Example 1, construct the pTEF-pADH1-nat-tTEF cassette, and obtain the plasmid Named pDH823.
[0064] 3. Perform AscI / XbaI double enzyme digestion on the pDH735 plasmid to obtain the attP1-ccdB-CAT-attP2 cassette, and subclone the attP1-ccdB-CAT-attP2 cassette into the plasmid pDH823 according to the method in Example 3 to construct pTEF-attP1 -ccdB-CAT-attP2-pADH1-nat-tTEF integration cassette, the obtained plasmid is pDH824, the full length is 6629bp.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com