lgr4 protein fragment and its application in the preparation of medicine for treating osteoclast-induced bone disease
A protein fragment, osteoclast technology, applied in the field of medicine for bone diseases, can solve the problems of high mortality, no literature report, developmental delay and other problems
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] Example 1: Purification and expression of LGR4 protein fragments
[0070] The LGR4 gene was amplified by PCR, and the amplified product was constructed on a PET-28a(+) prokaryotic expression vector, and PET-28a-ECD was transformed into Rosetta bacteria (full gold, cd801-03). The construction process refers to Table 1.
[0071] Table 1
[0072]
[0073]
[0074] According to the general prokaryotic expression method reported in "Acta Biophysica Sep. 2010, Vol. 26, No. 9: 790-798", the above vector was transformed into Rosetta bacteria to induce the expression of the LGR4 protein fragment. The experimental results are shown in Figure 1.
[0075] Figure 1a : LGR4 protein fragment (LGR4 ECD, h4) SDS-PAGE Coomassie Brilliant Blue result map, the arrow indicates the purified LGR protein fragment.
[0076] Figure 1b : Western blot results of LGR4 protein fragment (LGR4 ECD, h4), the target band in group h4 is the LGR4 protein fragment confirmed by His-tag antibody de...
Embodiment 2
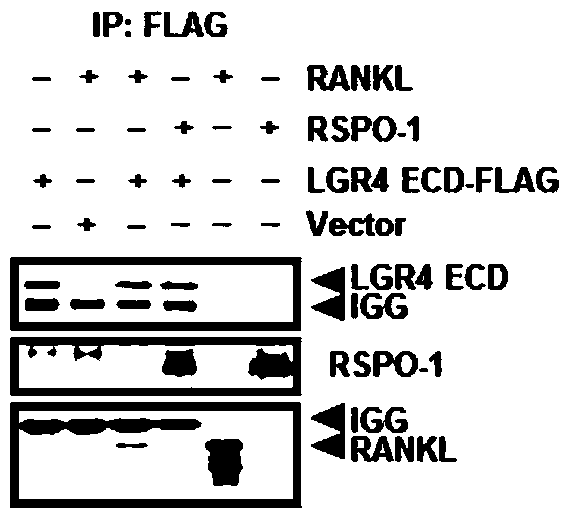
[0077] Example 2: LGR4 protein fragment binding RANKL experiment
[0078] Use lipofectamine2000 to overexpress the plasmid containing the above LGR4 fragment (see below for the construction process) in 293T cells according to the manufacturer's protocol. After 24 hours of transfection, the cells were washed once with PBS, scraped off with a cell scraper, and centrifuged at 12,000rpm for one minute. Lyse the cell block with RIPA weak lysate on ice for 15 minutes, centrifuge at 12000rpm at 4 degrees for 10 minutes, take the supernatant into a new EP tube, add 500ng RANKL and 200ngRSPO-1 (positive control) respectively; set the EP tube in a silent mixer Incubate overnight at 4°C (<12 hours), add 5 microliters of FLAG-M2 beads to each tube the next day, continue to incubate at 4°C for 3 hours, then centrifuge at 3000rpm at 4°C for 1 minute, remove the supernatant, and wash 3 times with PBS The beads were then lysed with 1XSDS loading buffer, boiled at 100°C for 10 minutes, and the...
Embodiment 3
[0091] Example 3: LGR4 protein fragment inhibits RANKL-RANK binding experiment
[0092] Use lipofectamine2000 to overexpress 10ug LGR4 fragment plasmids (see Table 2), 6ugRank, 4ug FLAG-Vector, and 2ugEGFP-N3 in 293T cells respectively according to the manufacturer's protocol. After 24 hours of transfection, the cells were washed once with PBS and washed with cell Scrape off with a scraper, centrifuge at 12000rpm for one minute, then lyse the cell mass with RIPA weak lysate on ice for 15 minutes, centrifuge at 12000rpm at 4 degrees Celsius for 10 minutes, take the supernatant into a new EP tube, and mix the protein lysate according to the following design: 2ugEGFP -N3+2ugFLAG, 2ugRANK+2ugFLAG, 2ugRANK+2ugLGR4 fragments, 2ugRANK+8ugLGR4 fragments, and then add 100ngRANKL respectively; EP tubes were incubated overnight at 4 degrees Celsius on a silent mixer, and 4 microliters of RANK antibody (1: 50), incubate at 4°C for 3 hours, then add 20 microliters of proteinA / G beads, cont...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com