Kit for detecting susceptibility to lung cancer and SNP (Single Nucleotide Polymorphism) marker thereof
A technology of susceptibility and markers, applied in the field of genetic testing, can solve the problems that the genetic markers of common diseases cannot be widely used, lack of methods for early detection of common diseases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0053] Example 1 is used to detect SNP sites related to the susceptibility of lung cancer
[0054] Among them, rs3842 is located in the gene ABCB1 region, rs212090 is located in the gene ABCC1 region, rs1926203 is located in the gene ACTA2 region, rs8034191 is located in the gene AGPHD1 region, rs2808630 is located in the gene APCS region, rs2273535 is located in the gene AURKA region, rs3117582 is located in the gene BAG6 region, rs7216064 is located in the gene BPTF region, rs2131877 is located in the gene C3orf21 region, rs3817963 is located in the gene C6orf10 region, rs1801270 is located in the gene CDKN1A region, rs1051730 is located in the gene CHRNA3 region, rs402710 and rs31489 are located in the gene CLPTM1L region, rs9981861 is located in the gene DSCAM region, rs1656402 is located in the gene EIF4E2 region, rs1656402 is located in the gene EIF4E2 region, rs12 rs1047840 is located in the gene EXO1 region, rs11080466 is located in the gene FAM38B region, rs4254535 is ...
Embodiment 2
[0055] Example 2 Detection Kit for Lung Cancer Susceptibility
[0056] 1. Preparation of the kit:
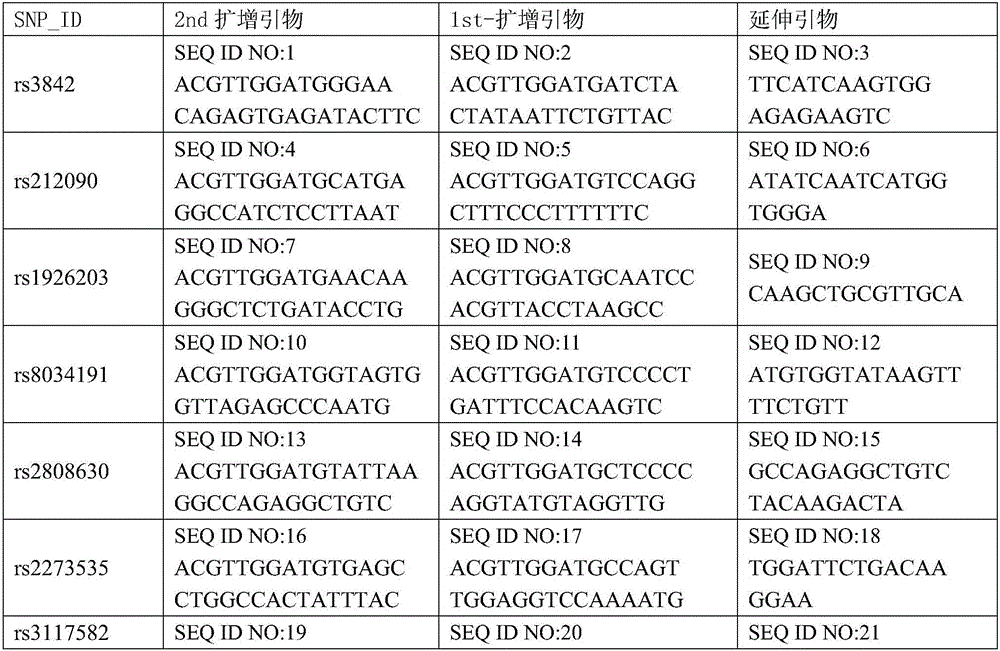
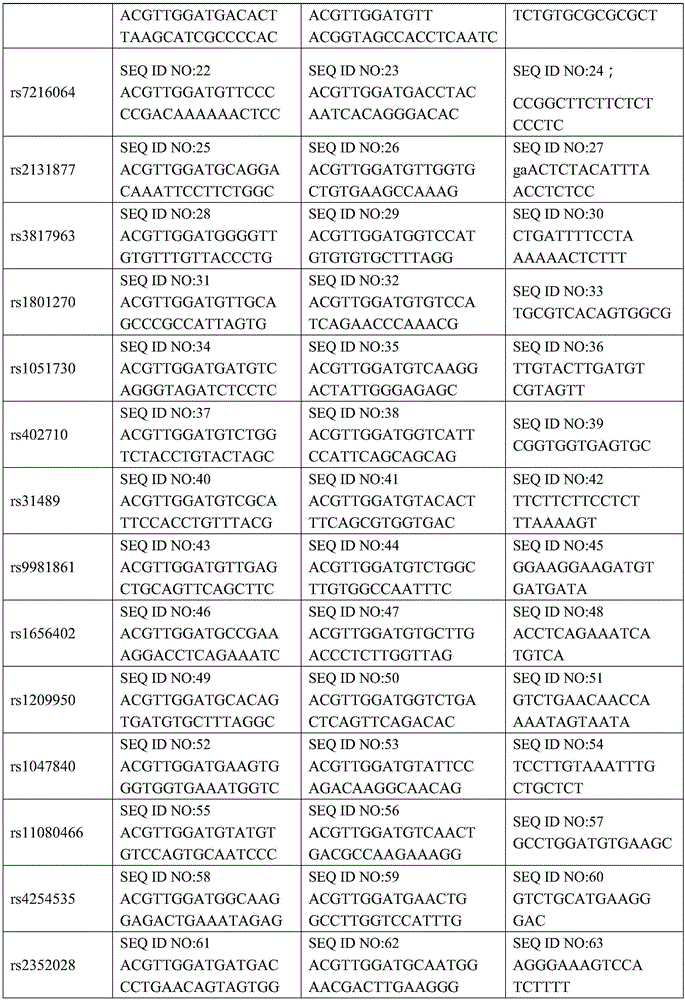
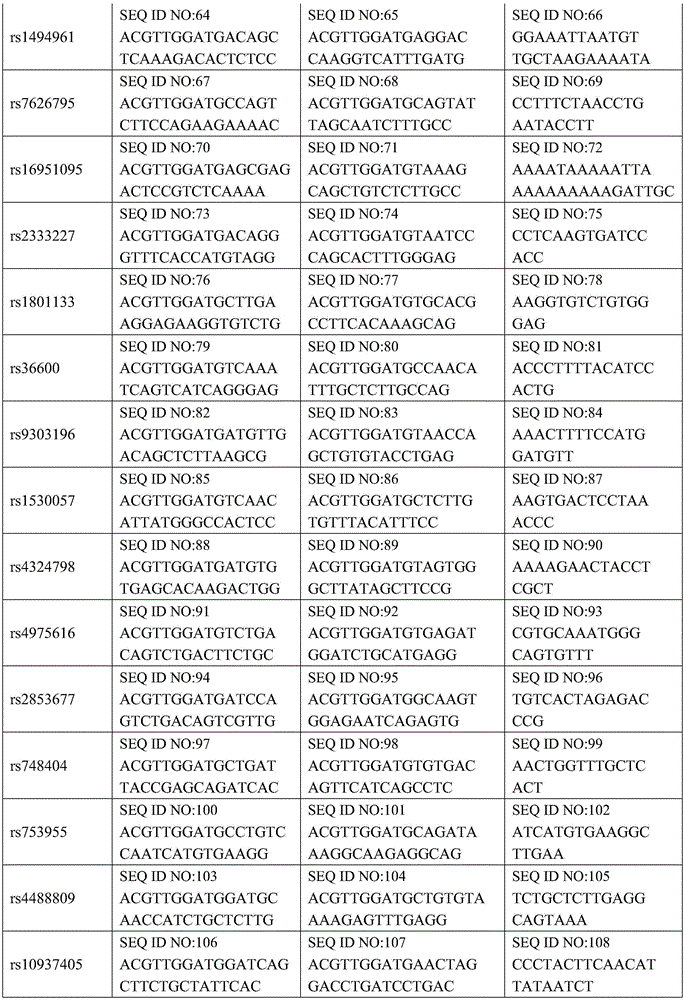
[0057] 1. Design and synthesize PCR amplification primers and single-base extension primers for the 36 SNP sites. The PCR amplification primers and single base extension primers of the SNP sites to be tested are shown in Table 1
[0058] Table 1 PCR amplification primers and single base extension primers of SNP sites to be tested
[0059]
[0060]
[0061]
[0062] 2. The kit also includes Taq enzyme, dNTP mixture, MgCl 2 Solution, PCR reaction buffer, enzyme digestion reaction buffer, deionized water.
[0063] 2. The detection method of the kit:
[0064] 1. Extraction of DNA
[0065] 1.1 DNA extraction from oral swab
[0066] 1) Put the oral swab in a 2ml centrifuge tube and add 400μl PBS.
[0067] 2) Add 20 μl QIAGEN Protease and 400 μl Buffer AL. Immediately vortex for 15s to mix. To ensure efficient lysis, sample and Buffer AL must be mixed immediately and ...
Embodiment 3
[0116] The literature cited by 36 SNP sites closely related to lung cancer in Example 3 is shown in Table 5
[0117] Table 5 Literature cited by 36 SNP sites closely related to lung cancer
[0118]
[0119]
[0120]
[0121]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com