Preparing and identification method for chrysanthemum CmACO gene monoclonal antibodies
A technology of monoclonal antibody and chrysanthemum, applied in the field of bioengineering, can solve the problems of key enzyme preparation and functional blank
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0046] Provided is a method for preparing a chrysanthemum CmACO gene monoclonal antibody, comprising the following steps:
[0047] Step 1, preparing and sequencing the chrysanthemum CmACO gene;
[0048] Step 2, finding the antigenic fragment from the amino acid sequence of the chrysanthemum CmACO gene, and synthesizing the chrysanthemum CmACO antigenic protein;
[0049] Step 3: immunize experimental animals with the chrysanthemum CmACO antigen protein, and take animal cells to fuse with myeloma cells and coat with antigens to obtain hybridoma cell lines.
[0050] Step 4: The hybridoma cell line is proliferated, purified and screened to obtain the monoclonal antibody of the chrysanthemum CmACO gene.
[0051] What needs to be understood is that a gene (genetic element) is a DNA segment that has a genetic effect (some viruses such as tobacco mosaic virus and HIV have genetic material that is RNA). Genes support the basic structure and performance of life. It stores all the inf...
Embodiment
[0106] 1. According to the chrysanthemum EST database information, design a pair of primers:
[0107] CmACOF: GAACCTGAGAACAA TGGAAACCTT
[0108] CmACOR: CAAGACACCAGACCGATAAT
[0109] The target fragment was obtained by RT-PCR and sequenced.
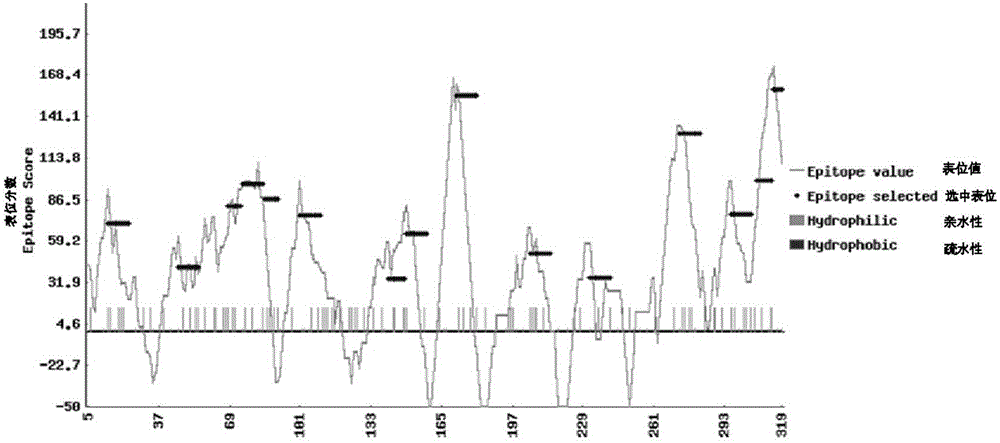
[0110] 2. Using the amino acid sequence translated by CmACO ORF as the material, various methods in DNAStar were used to predict the epitope of the antigen cell.
[0111] The epitope map can comprehensively display the epitope index of each region in the target protein and the selected epitope, and the X-axis is the starting position of each 10 peptides in the target protein. The Y axis is the epitope index score. The higher the score, the more likely the 10-peptide is a successful epitope. The gray curves are the score curves for each 10 peptides. Blue indicates the selected 10-peptide fragments. The short bars on the X-axis represent the distribution of hydrophilicity in the target protein: green represents hydrophilicity, and red...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com