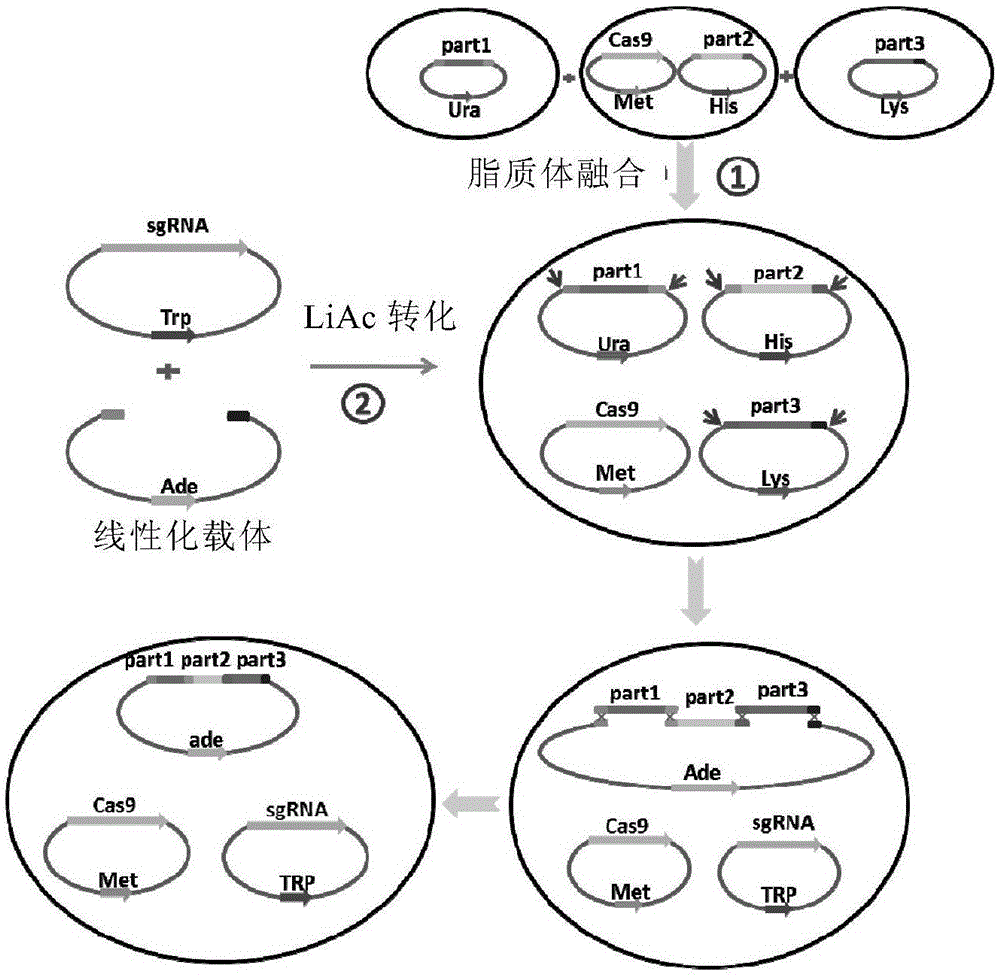
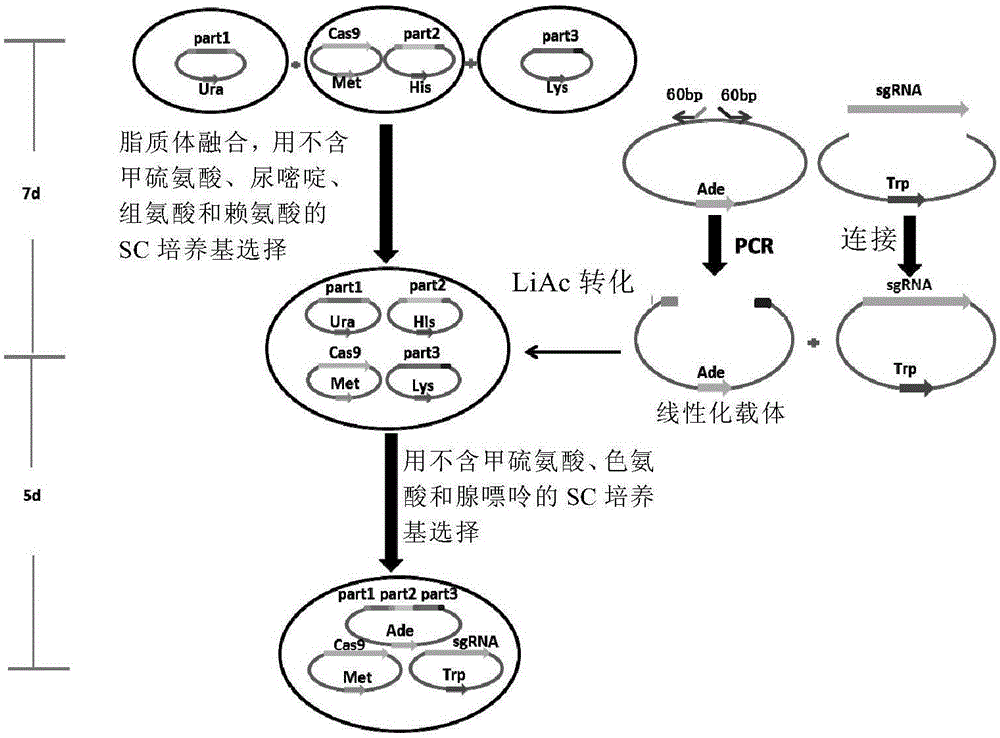
CRISPR/Cas9-mediated large DNA fragment assembling method
A technology for nucleic acid constructs and yeast, applied in recombinant DNA technology, fungi, bacteria, etc., can solve problems such as difficult operation and low efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0066] Example 1 : Splicing of two large fragments of DNA
[0067] In this example, the pSP5 and pTP3-U plasmids constructed in Saccharomyces cerevisiae VL6-48 in our laboratory were selected (Table 2). pSP5 contains 116643bp of donor DNA, named SP5; pTP3-U contains 184475bp of donor DNA, named TP3. Both SP5 and TP3 sequences are derived from Escherichia coli, which are large plasmids obtained by amplifying certain essential genes of Escherichia coli by PCR and splicing two or three levels in yeast. The right end of SP5 and the left end of TP3 have the same 491bp source sequence.
[0068] Table 2: Plasmid pSP5, pTP3-U information
[0069] Donor plasmid filter marker Donor DNA name Insert size (bp) pSP5 HIS3 SP5 116643
[0070] pTP3-U URA3 TP3 184475
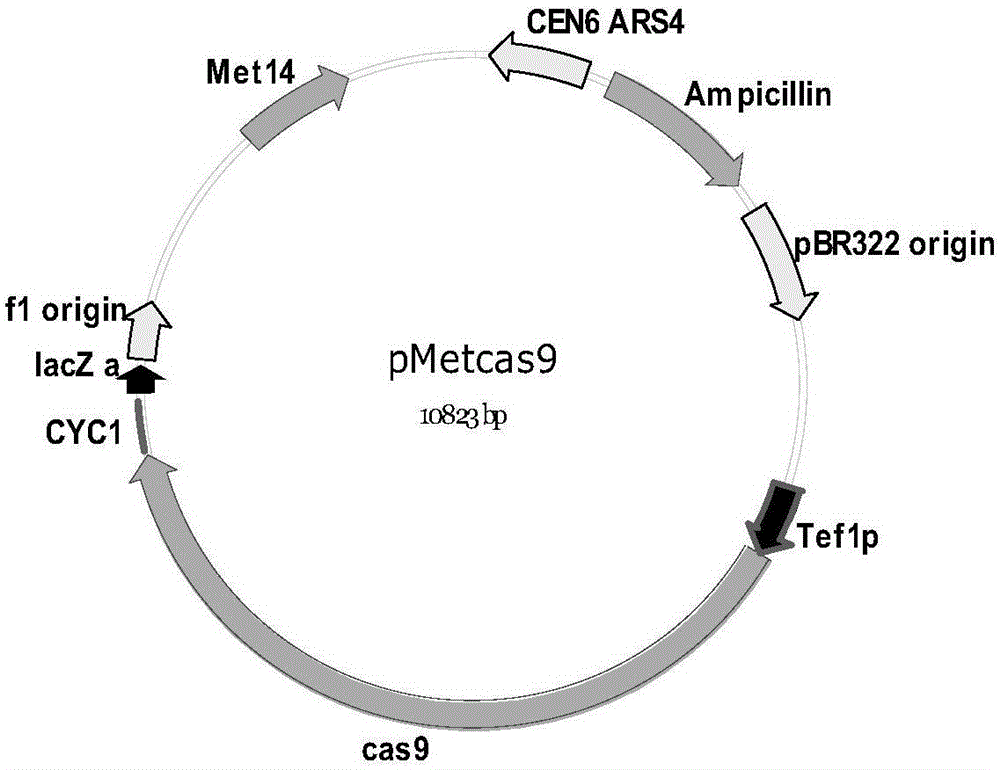
[0071] The Cas9 expression plasmid pMetcas9 ( image 3 , SEQ ID NO: 1) was transferred into the yeast cell VL6-48 containing the pSP5 plasmid. Protoplast fusion of VL6-48 / p...
Embodiment 2
[0120] Example 2 : Splicing of three large fragments of DNA
[0121] In this example, the pTP1, pTP2 and pTP3-L plasmids constructed in Saccharomyces cerevisiae VL6-48 in our laboratory were selected (Table 4).
[0122] Table 4: Plasmid pTP1, pTP2, pTP3-L information
[0123] filter marker insert clip name Insert size (bp) pTP1 HIS3 TP1 177147 pTP2 URA3 TP2 297952 pTP3-L LYS2 TP3 184475
[0124] pTP1 contains 177147bp of donor DNA, named TP1; pTP2 contains 297952bp of donor DNA, named TP2; pTP3-L contains 184475bp of donor DNA, named TP3. The sequences of TP1, TP2 and TP3 are all derived from Escherichia coli, and their construction methods are similar to those of pSP5 and pTP3-U in Example 1. There is a 385bp homologous sequence between the right end of TP1 and the left end of TP2, and a 491bp homologous sequence between the right end of TP2 and the left end of TP3. The Cas9 expression plasmid pMetcas9 (see image 3 and ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com