Genetic toxicant detection vector and detection method
A genotoxicity and detection carrier technology, applied in the field of genotoxic substance detection, can solve the problems of long detection time, cumbersome steps, difficult preservation of strains, etc., and achieves the effects of simple and easy operation, high detection sensitivity, and no risk of disease.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
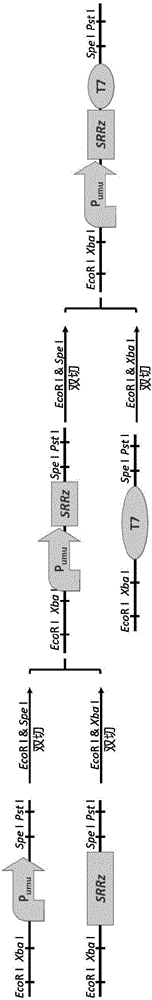
[0047] Construction of genotoxic response vector
[0048] Using pUC18 as the starting vector, the genotoxic response vector pUST was constructed. The specific construction method is as follows:
[0049] a) Synthetic Genetic Response Promoter P umu (SEQ ID No. 1) and T7 terminator sequences, EcoRI and XbaI sites were added upstream of the promoter and T7 terminator, and SpeI and PstI restriction sites were added downstream.
[0050] b) Using lambda phage genomic DNA as a template, amplify the SRRz gene. EcoRI and XbaI, SpeI and PstI restriction sites were also added upstream and downstream of the SRRz gene.
[0051] c) Using the Biobricks method, using enzyme cleavage and ligation methods, each element is connected in the sequence of promoter-cleavage gene-terminator, and inserted into the pUC18 vector to obtain the genotoxic response vector pUST.
[0052] d) Transfer the vector pUST into E.coli BL21 competent cells to obtain recombinant E.coli BL21 / pUST for the detection o...
Embodiment 2
[0054] Calculation of cracking efficiency
[0055] 0.5h after the Escherichia coli detection solution was in contact with the sample to be tested, the light absorption value (OD) of each test bacterial solution at 600 nm was determined respectively. 600 ).
[0056] Cracking rate (%)=(A-B) / A*100%
[0057] Wherein, A—the bacterial liquid OD of adding methanol 600
[0058] B—the OD of the bacterial liquid added to the test sample 600 .
Embodiment 3
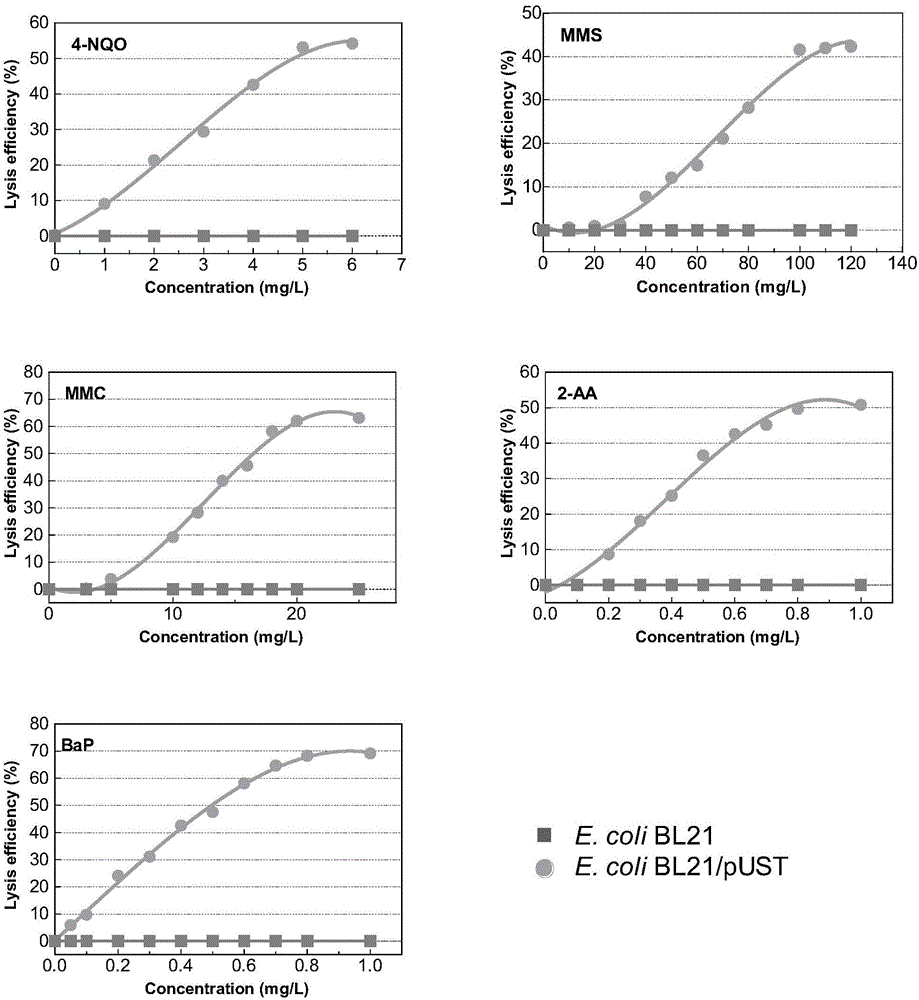
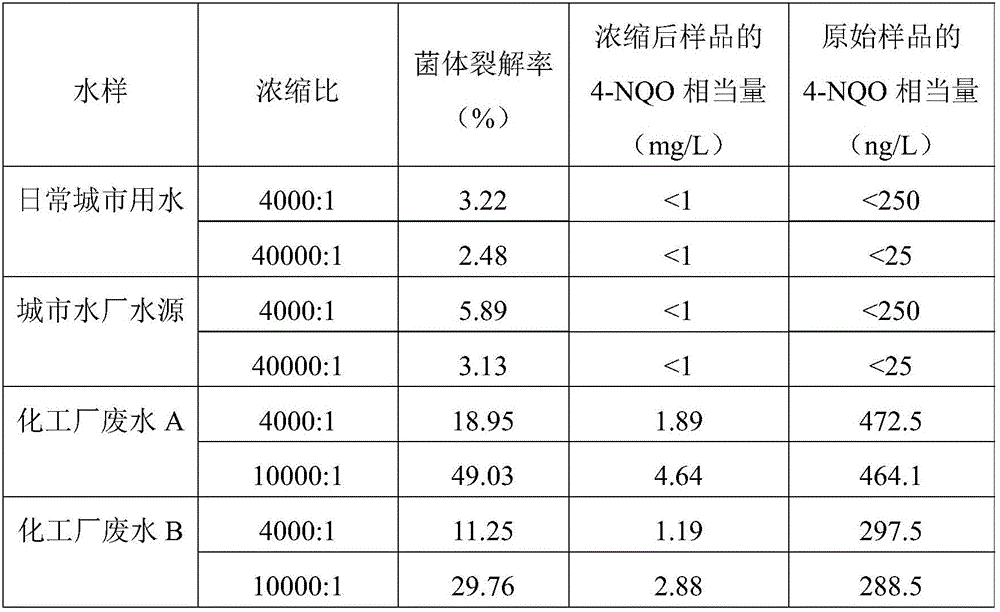
[0060] Genotoxic Substance Detection
[0061] 1. Recovery activation of recombinant E. coli
[0062] (1) The wild-type Escherichia coli (E.coli BL21 / pUC18) and the recombinant bacteria (E.coli BL21 / pUST) were streaked into LB plate medium from a -80° refrigerator, and the culture was resumed at 37°C. 14h.
[0063] (2) Pick a single colony, inoculate it into LB medium, and culture at 37°C and 250rpm for 12-16h.
[0064] 2. Preparation of Escherichia coli detection solution
[0065] Inoculate the revitalized bacterial solution into fresh LB medium at a volume ratio of 1:100, add 190 μl volume to a 96-well culture plate, and cultivate to OD at 35-37 °C and 800 rpm. 600 to 0.15-0.25.
[0066] 3. Contact with the test sample
[0067] Methanol and 10 μl each of 4-NQO, MMS, MMC, 2-AA and BaP with different concentrations were added to the 96-well plate respectively, and cultured at 35-37 °C and 800 rpm for 0.5 h, and 3 parallel groups were set up.
[0068] 4. The experimental r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com