Apparent modification detection primer pair based on ATP4B genosome DNA and kit thereof
A gene body and kit technology, applied in the field of gene body DNA methylation modification detection, can solve problems such as inability to up-regulate
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
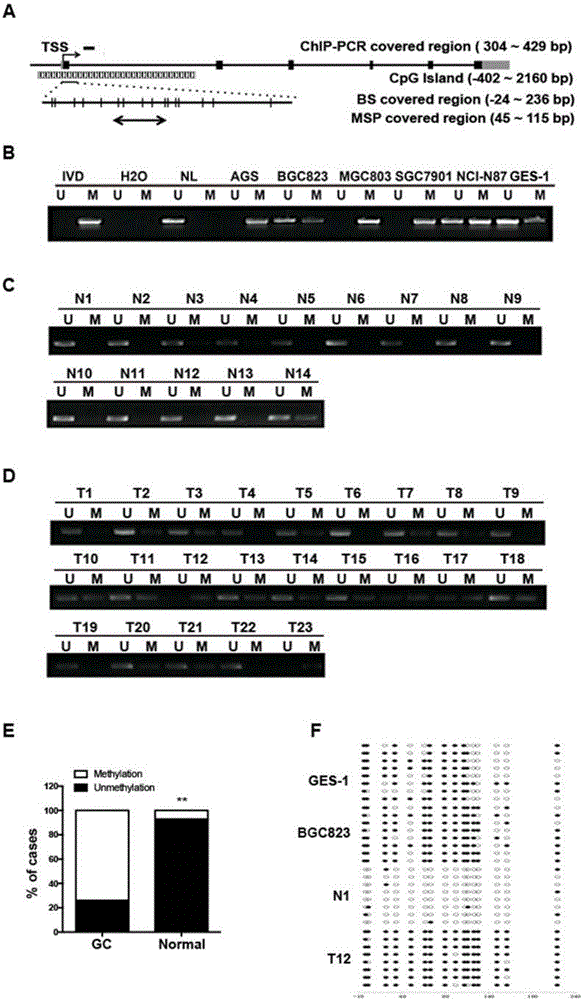
[0039] Example 1: Design of special primers for ATP4B gene DNA methylation
[0040] According to the distribution characteristics of DNA methylation modification in the genome (Maunakea AK., et al. Nature 2010, 466(7303); Jjingo D., et al. Oncotarget 2012; 3(4).), through manual comparison analysis, Select the DNA region rich in CG in the specific ATP4B gene sequence from GenBank, design multiple primer pairs with the software Primer Express 3.0, and screen out the following methylated primers and non-methylated primers. All primers are provided by Tianyi Huiyuan Technology Company synthesis.
[0041] The nucleotide sequence of the upstream primer of the methylated primer is: 5'-GAGTTTTAGCGTTATTGTTGGAATTCGGATAC-3' (shown in SEQ ID No. 3), and the nucleotide sequence of the downstream primer is: 5'-GTACCCCACCGAAACAAAATACG-3' (such as SEQ ID No. 4); the nucleotide sequence of the upstream primer of the unmethylated primer is: 5'-GGAGTTTTAGTGTTATTGTTGGAATTTGGATATG-3' (as shown in SEQ...
Embodiment 2
[0042] Example 2: Composition of a kit for detecting ATP4B gene DNA methylation modification
[0043] A kit for detecting ATP4B gene DNA methylation modification, that is, a kit for early diagnosis of gastric cancer, detection of chemotherapy sensitivity and detection of drug targets, including nucleotide sequences such as SEQ ID No. 3 and SEQ ID The methylated primer shown in No.4, the nucleotide sequence of the unmethylated primer shown in SEQ ID No.5 and SEQ ID No.6, the methylated control DNA template for MSP (modified by sulfurization) Afterwards, more than 90% hypermethylated DNA appears in the ATP4B gene body region), unmethylated control DNA template (ATP4B gene body region shows unmethylated normal tissue cell DNA), deionized water as a negative system control, PCR Reaction reagents.
Embodiment 3
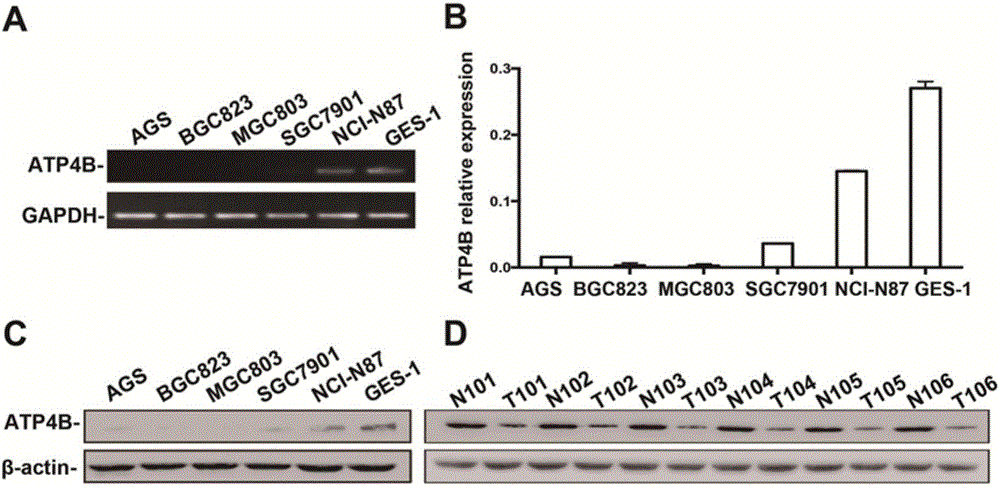
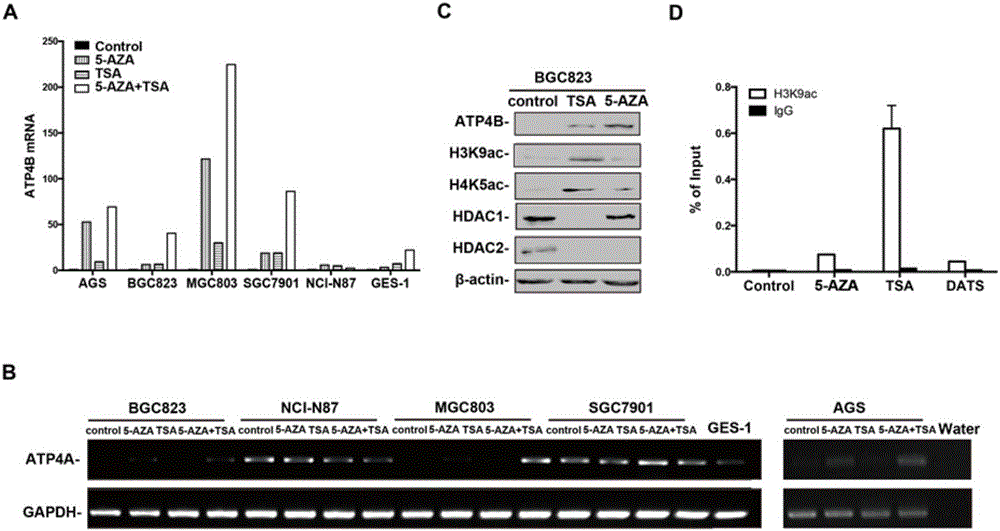
[0044] Example 3: Detection of ATP4B methylation in cells and tissues
[0045] (1) The extraction process of cell line DNA
[0046] The gastric cancer cell lines and normal gastric cell lines in good growth condition and in the logarithmic growth phase were digested with trypsin and then counted, and collected in a 1.5mL centrifuge tube. The number of cells was about 10 5 To 10 6 One. Centrifuge at 13000 rpm for 10 seconds, discard the supernatant and add 200 μL 1×PBS to resuspend the cells. After washing again, resuspend the cells in 180 μL 1×PBS. The following steps refer to the Biomed Genomic DNA Rapid Extraction Kit Operation Manual. The specific operations are as follows:
[0047] Add 20 μL of 20 mg / mL proteinase K to the cell suspension, mix well, add 200 μL of binding solution, and place at 70°C for 10 minutes. After cooling, add 100 μL of isopropanol, add the mixture to the adsorption column, centrifuge at 13000 rpm for 30 seconds, and discard the waste liquid. Add 500μL of...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com