Protein rapid detection method based on matched microalgae protein characteristics sequence label and system thereof
A protein sequence and characteristic sequence technology, applied in proteomics, sequence analysis, genomics, etc., can solve the problems of time-consuming data sets, inability to provide all sequence information, and misleading users
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
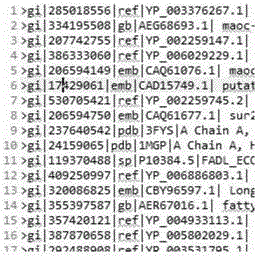
[0115] For the input of characteristic enzyme parameters, take the following data as an example:
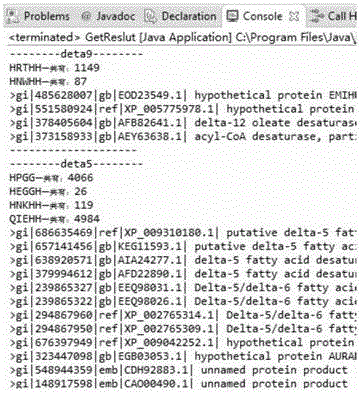
[0116] The fatty acid desaturase family is mainly composed of six desaturases, delta-4, delta-5, delta-6, delta-8, delta-9 and delta-12, and each fatty acid desaturase sequence contains multiple specific proteins feature fragment. For example, the following desaturases need to be searched:
[0117] Delta-4: HPGG, HMGGH, HNKHH, QIEHH
[0118] Delta-5: HPGG, HEGGH, HNKHH, QIEHH
[0119] Delta-6: HDTLH, HNLHH, QIEHH
[0120] Delta-8: HPGG, HDYLH, HNTHH, QTEHH
[0121] Delta-9: HRTHH, HNWHH
[0122] Delta-12: HECGH, HAKHH, HVVHH
[0123] i∈1-15, the protein characteristic value fragment (Protein Character Fragment) j is defined as:
[0124] PCFi∈[HRTHH,HNWHH,HPGG,HEGGH,HNKHH,QIEHH,DHTLH,HNLHH,HECGH,HAKHH,HVVHH,HDYLH,HNTHH,QTEHH,HMGGH]
[0125] For step 1, the obtained text files to be retrieved are often hundreds of megabytes or even several gigabytes. To find delta sequences...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com