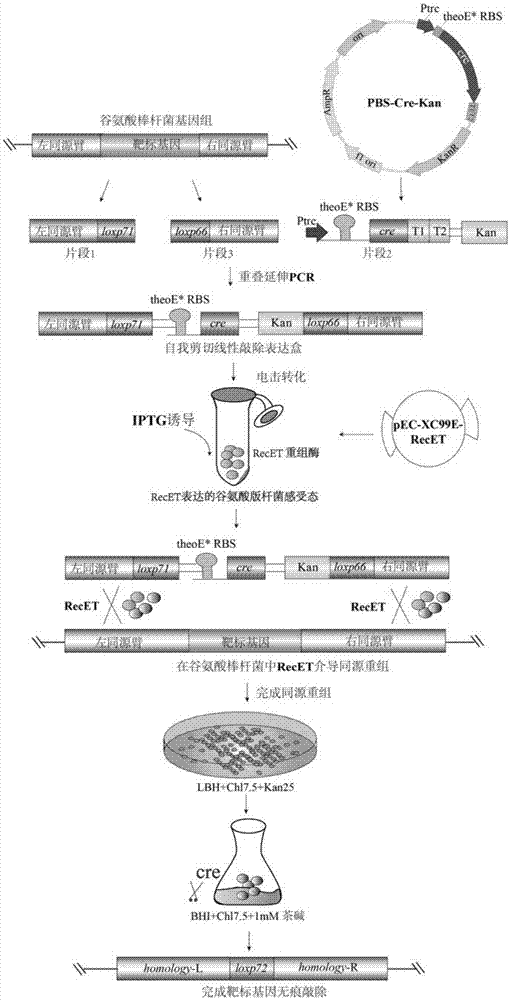
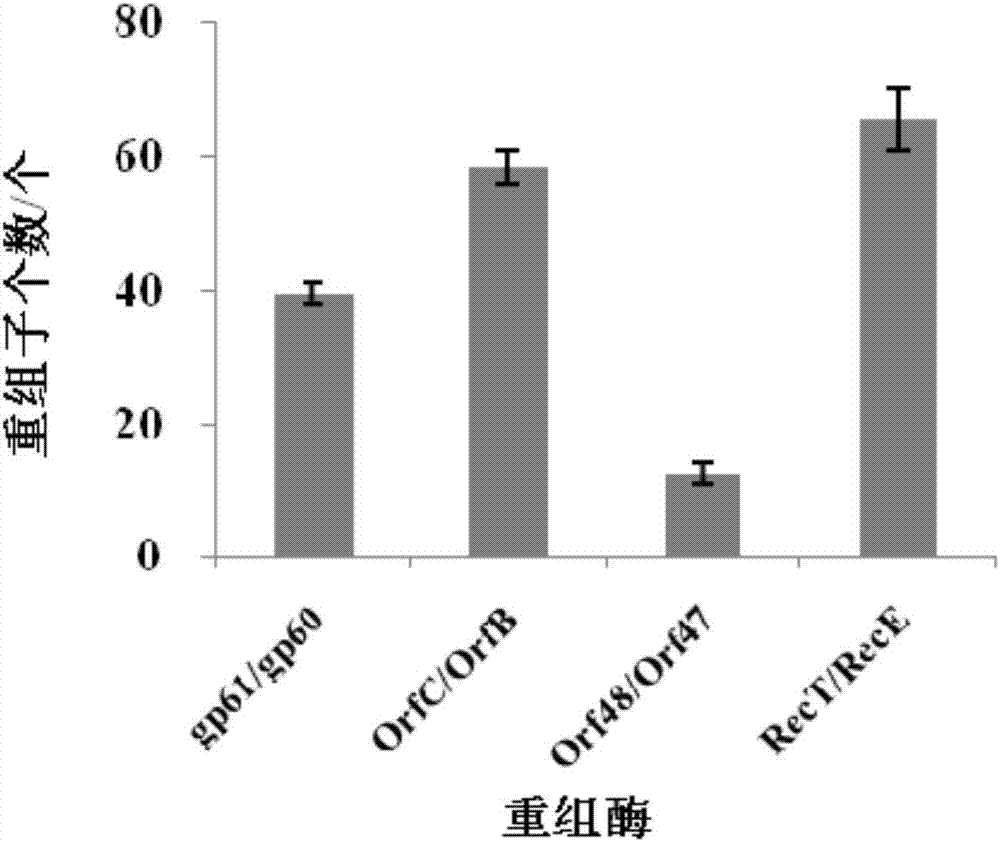
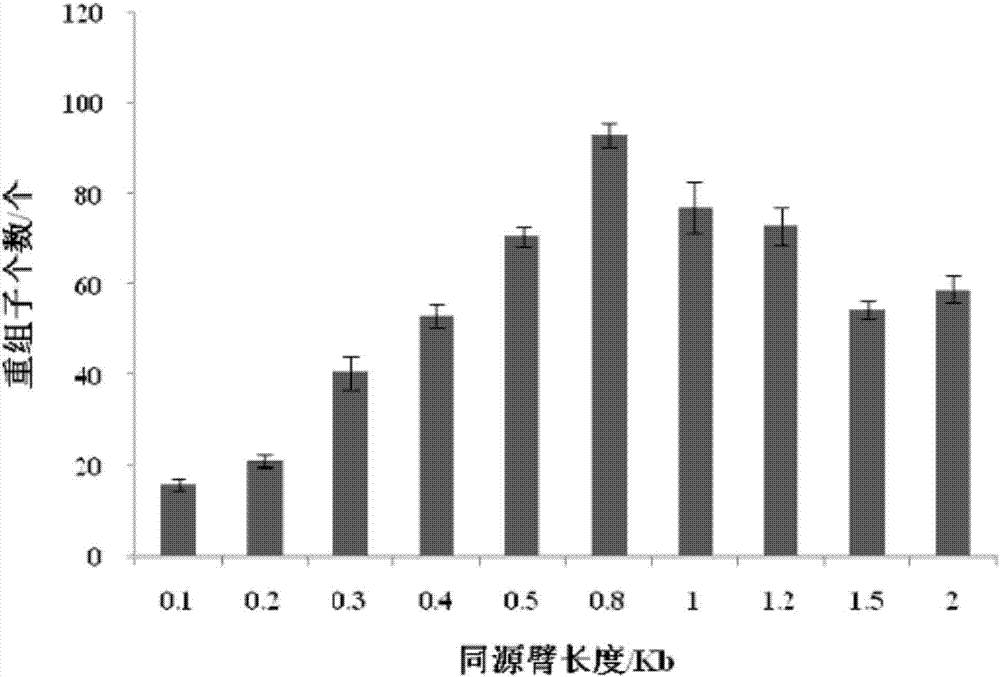
Continuous traceless gene knockout method for corynebacterium glutamicum
A Corynebacterium glutamicum, traceless knockout technology, applied in the field of genetic engineering, can solve the problems of cumbersome and lengthy knockout process and low homologous recombination, and achieve the effect of simple and effective screening process and time saving.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0047] Example 1 Exonuclease-recombinase to the recombination of linear double-stranded DNA in Corynebacterium glutamicum
[0048] 1. Preparation of Corynebacterium glutamicum Competent Cells Expressed by Exonuclease-Recombinase
[0049] Design primers recT / recE-S: 5′-CAAAAAGGAGGCCCTTCAGATGAGCACAAAACCACTCTTC-3′ and recT / recE-A: 5′-GGCACCAATAACTGCCTTAATTATTCCTCTGAATTATCGA-3′ to amplify the gene recT+recE;
[0050] Design primers orfC / orfB-S: 5′-CAAAAAGGAGGCCCTTCAGATGACAAAATTATGTTTTAGTG-3′, orfC / orfB-A: 5′-GGCACCAATAACTGCCTTAATTAAGCCTTTATCCTGATTAGT-3′, amplify gene orfC+orfB;
[0051] Design primers orf48 / orf47-S: 5′-CAAAAAGGAGGCCCTTCAGATGGCTATTGCAAAAGAAAAGAC-3′, orf48 / orf47-A: 5′-GGCACCAATAACTGCCTTAATTAGATCATTGACCCTTGAACC-3′, amplify orf48+orf47 gene;
[0052] Design primers gp60 / 61-S: 5′-ACAAAAAGGAGGCCCTTCAGATGAGTGTGCCCACACAGGACGGA-3′, gp60 / 61-A: 5′-GGCACCAATAACTGCCTTAATCATGCGTTGGGCCCGTCGAACAT-3′, amplify gp60+gp61 gene;
[0053] Design primers PEC-A: 5'-CTGAAGGGCCTCCTTTTTGT...
Embodiment 2
[0066] Example 2 Obtaining of single gene traceless knockout strain ATCC14067-RecET-MargR
[0067] 1. Preparation of self-cut linear double-stranded DNA knockout expression cassette ΔArgR
[0068] Design primers PBS-Kan-S: 5'-TCGATCCTTTTTAACCCATTGCAGGAATTCGATATCAAG-3', PBS-Cre-A: 5'-GCGACACGAATTATGCAGTTTGTTATCCGCTCACAATTC-3', amplify the 2875bp plasmid pBluescript II SK (+) backbone fragment;
[0069] Primers theoE-Cre-S: 5′-ACTGCATAATTCGTGTCGCTCAAG-3′, theoE-Cre-A: 5′-CTTTGCGCTTGCGCGGAATTAATTCATGAGCG-3′ were designed to amplify the 1594bp inducible Cre enzyme expression cassette;
[0070] Design primer Kan-S: 5′-AATTAATTCCGCGCAAGCGCAAAGAGAAAGCA-3′,
[0071] Kan-A: 5′-ATGGGTTAAAAAGGATCGATCCTC-3′, amplified 1074bp Kan resistance screening marker expression cassette;
[0072] After recovering the above three fragment gels, they were assembled with the NEBuilder HiFi DNA Assembly Master Mix (NewEngland BioLabs) kit to obtain the universal plasmid PBS-Cre-Kan.
[0073] Universa...
Embodiment 3
[0080] Example 3 Superimposed traceless knockout of the crtB gene in the traceless knockout strain ATCC14067-RecET-MargR
[0081] Design primers crtB-L-S (1) according to the method in Example 2: 5'-GAAGTGTTGATAGAAATGACCTCA-3', crtB-L-loxp71: 5'-TGCAGTATAACTTCGTATAATGTATGCTATACGAACGGTAATGAAGACATCAACTACAACTCC-3', amplify 840 bp containing crtB using the genome of ATCC14067 as a template The DNA sequence of the left homology arm of the gene and the loxp71 sequence (C-1);
[0082] Design primers crtB-R-loxp66: 5′-ACCCATATAACTTCGTATAGCATACATTATACGAACGGTATCATAGCTGAGCCTGCTTCTGG-3′, crtB-R-A(1): 5′-GTCACTAGTGCTGACTCCCCCTCT-3′, amplify 850bp right homology arm containing loxp66 and crtB gene using ATCC14067 genome as template DNA sequence (C-3).
[0083] Using crtB-L-S(1), crtB-R-A(1) as primers, fragment C-1, 2712bp universal Cre-Kan expression cassette (fragment 2) and fragment C-3 as template, fusion 4322bp ΔCrtB self-cleavage A linear knockout expression cassette for .
[0084] A...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com