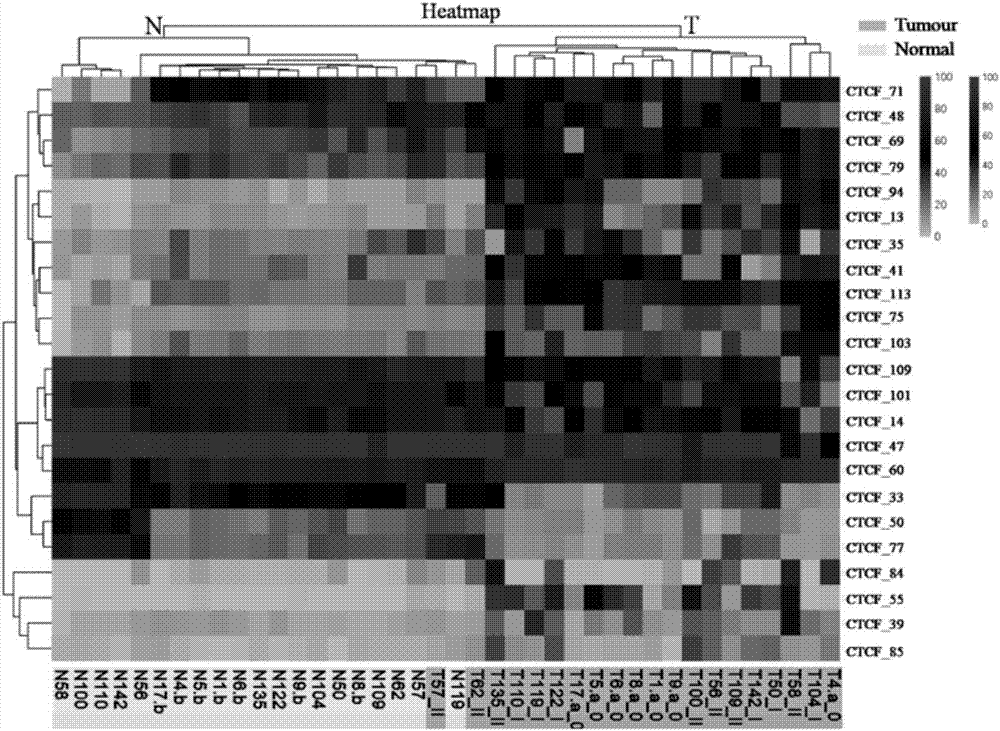
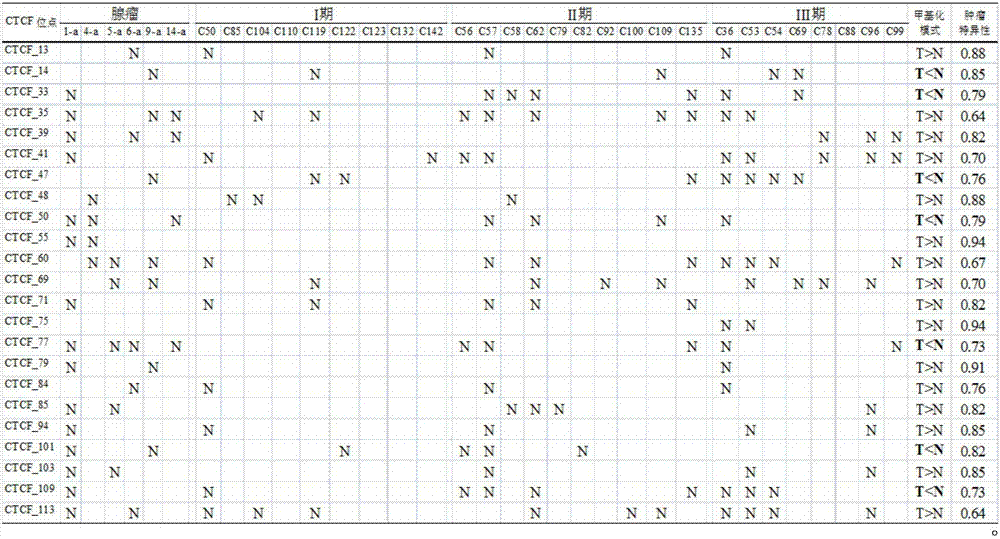
Application of DNA binding site CTCF_55 of multifunctional transcriptional regulation factor CTCF
A technology of transcription regulator and binding site, which is applied in the application field of the DNA binding site CTCF_55 of the multifunctional transcription regulator CTCF
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0023] Example 1: Screening of the DNA Binding Site of the Multifunctional Transcription Regulator CTCF
[0024] 1. Sample collection
[0025] From April 2014 to August 2016, 187 cases of fresh tumor tissues and 84 cases of corresponding normal tissues (more than 6cm from the tumor edge) were collected from the First Affiliated Hospital of Kunming Medical University from April 2014 to August 2016; 108 cases of adenoma tissue. A total of 379 samples were taken, including 84 cases of normal tissues and 295 cases of tumor tissues; the clinical data of the above samples are shown in Table 1:
[0026] Table 1 Sample information table
[0027] .
[0028] 2. Genomic DNA extraction and bisulfite conversion
[0029] Genomic DNA was extracted using the QIAamp DNA Mini Kit (Product No. 51304) kit from QIAGEN, and all operations were performed according to the instructions of the kit. Bisulfite conversion was performed using the EpiTect Fast DNA Bisulfite Kit (Catalog No. 59826) fr...
Embodiment 2
[0056] Example 2: Enlarging the sample size for further screening of 10 loci
[0057] In order to improve the specificity and sensitivity of the screened molecular markers in the detection of colorectal adenoma, we added 50 colorectal adenoma samples to further screen the 10 CTCF binding sites screened in Example 1 Adding the mass spectrometry data of 40 samples in Example 1, this step of screening included a total of 90 samples, of which 20 were normal controls, 57 were adenomas, 6 were stage I tumors and 7 were stage II tumors. Table 6 shows the performance of these 10 CTCF binding sites in distinguishing tumor tissue from normal tissue. It can be seen from Table 6 that these 10 CTCF binding sites all have a strong ability to distinguish tumor tissue from normal tissue (AUC ≥ 0.85). Their sensitivities ranged from 44.64% to 88.89% at a specificity of 95%. Our goal is to establish an early diagnosis technology for colorectal tumors that includes as few molecular markers as ...
Embodiment 3
[0060] Example 3: Verify the detection accuracy of the CTCF binding site CTCF_55 of the present application in a large sample
[0061] In order to verify the detection accuracy of the binding site CTCF_55 to be protected in this application, we selected 379 colorectal samples (Table 1), including 84 normal samples and 295 tumor samples (108 adenomas, 39 stage I tumors) , 101 cases of stage II tumors and 47 cases of stage III tumors), continue to use DNA mass spectrometry to detect the DNA methylation level of the binding site in 379 cases of colorectal samples, the detection method is the same as in Example 1 "DNA mass spectrometry DNA methylation analysis" has the same content, using the primer pair shown in SEQ ID NO:2 and SEQ ID NO:3.
[0062] According to the results of DNA mass spectrometry, we statistically analyzed the sensitivity of CTCF_55 in the detection of adenoma, stage I tumor, stage II tumor, stage III tumor and all tumors when the AUC value and specificity were...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com