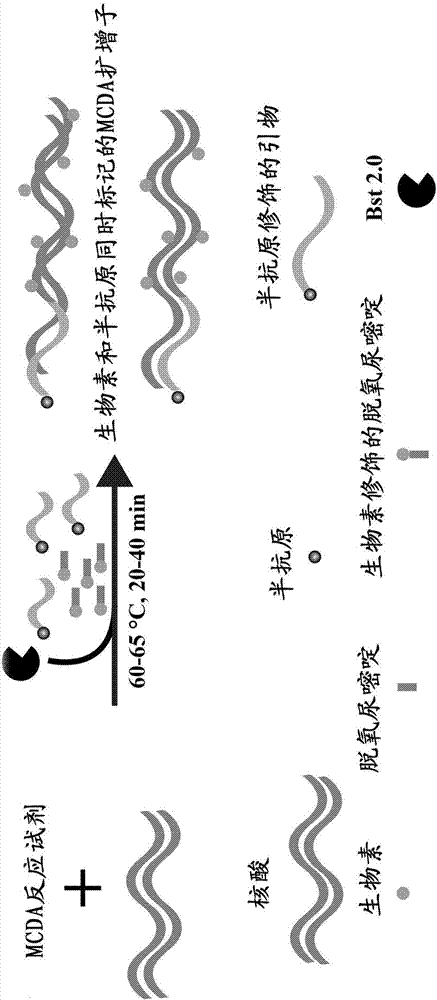
AUDG (antarctic thermal sensitive uracil deoxyribonucleic acid glycosylase) mediated multiple cross displacement amplification and biosensing combined nucleic acid testing technique
A biosensing and biosensor technology, applied in the field of microorganisms and molecular biology, can solve problems such as cross-contamination, and achieve the effects of eliminating pollutants, high sensitivity, and convenient methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Embodiment 1.MCDA amplification
[0061] 1. Construction of detectable products by MCDA amplification
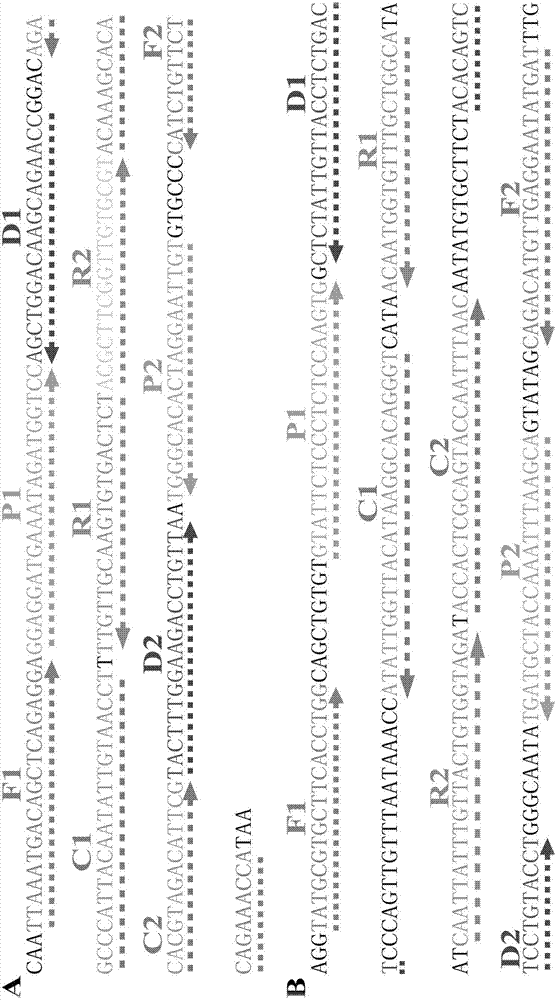
[0062] The MCDA reaction system includes 10 primers, which recognize 10 regions of the target sequence, including 2 internal cross primers, namely CP1 and CP2 (Cross Primer, CP), 2 displacement primers (Displacement Primer), namely F1 and F2, and 6 Amplification primers, namely D1, C1, R1, D2, C2 and R2. In order to construct a detectable product, select any one of the 10 primers, label the hapten (fluorescein or digoxin) at the 5' end, and the newly labeled primers are named F1*, F2*, CP1*, CP2*, C1*, C2*, D1*, D2*, R1* and R2*. In the present invention, CP1* is taken as an example to illustrate the principle of the present invention.
[0063] Under the given constant temperature conditions, the double-stranded DNA is in a dynamic equilibrium state of half-dissociation and half-binding. When any primer performs base pairing extension to the complementary part of t...
Embodiment 2
[0073] Embodiment 2. measure the optimal reaction temperature of MCDA technology
[0074] Under standard reaction system conditions, add the corresponding MCDA primers designed for HPV16 and HPV18 DNA templates, and the template concentration is 5×10 4 copies / µl. The reaction was carried out under constant temperature conditions (60-67°C), and the results were detected by a real-time turbidimeter, and different dynamic curves were obtained at different temperatures, see Figure 7 with Figure 8 . 61-64°C is recommended as the optimal reaction temperature for the two sets of MCDA primers. In the follow-up verification of the present invention, 63° C. was selected as the constant temperature condition for MCDA amplification. Figure 7 Represents the temperature dynamic curve of the MCDA primers designed for the detection of HPV16 against the E7 gene; Figure 8 It shows the temperature dynamic curve of the MCDA primer designed for detecting HPV18 aimed at the L1 gene.
Embodiment 3
[0075] Embodiment 3.AUDG-MCDA-LFB detects the sensitivity of single target
[0076] Use the serially diluted plasmid (pMD18-T-HPV16: 5×10 5 , 5×10 4 , 5×10 3 , 5×10 2 , 5×10 1, 5×10 0 and 5×10 -1 copy / microliter) after performing standard MCDA amplification reaction, the result is displayed by LFB detection. For HPV16 detection, the detection range of MCDA-LFB is 5×10 5 ~5×10 0 copies / microliter, LFB appears red lines in TL1 and CL regions ( Figure 9 1-6 in A). When the amount of genome template in the reaction system is reduced to 5×10 0 When copying below, LFB only appears a red line in the CL area, indicating a negative result ( Figure 9 7-8 in A). Figure 9 A is to use LFB to visualize and read the MCDA amplification results: Figure 9 1 to 6 in A indicate that the template amount of pMD18-T-HPV16 is 5×10 5 , 5×10 4 , 5×10 3 , 5×10 2 , 5×10 1 , 5×10 0 copies / µl, Figure 9 7-8 in A respectively represent that the template amount of pMD18-T-HPV16 is 5×1...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com