Method for improving activity of rate-limiting enzyme in methanol metabolism process of escherichia coli
A technology of methanol metabolism pathway and Escherichia coli, which is applied in the field of assembly and regulation of Escherichia coli methanol metabolism pathway, can solve the problems of low utilization efficiency, low efficiency and backwardness of strain genetic manipulation, etc., and achieve the effect of realizing methanol metabolism and improving the effect of methanol metabolism
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] Example 1: Escherichia coli Methanol Metabolic Pathway Assembly
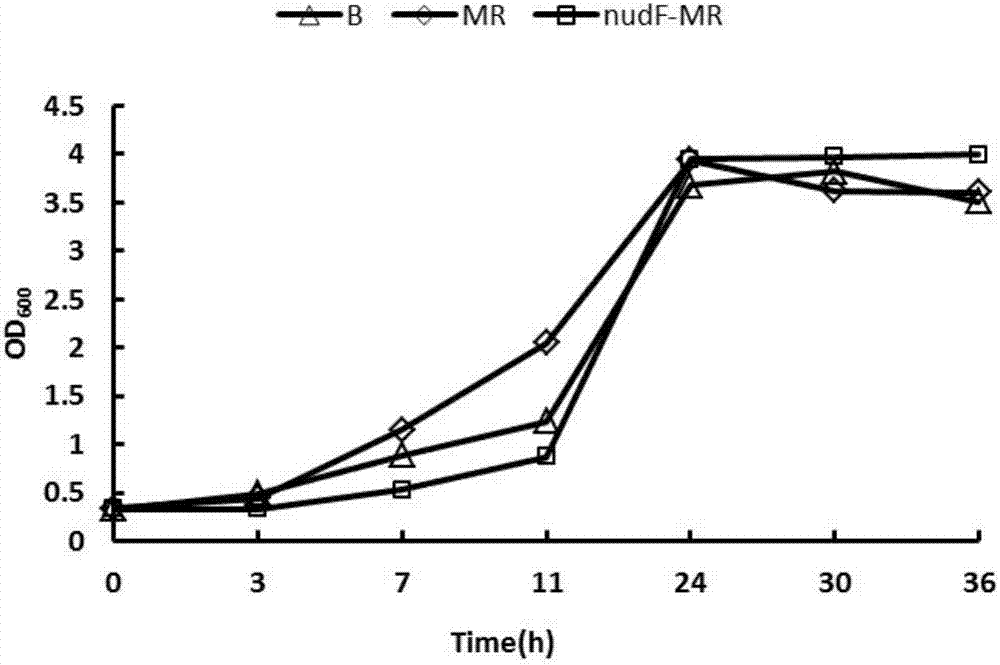
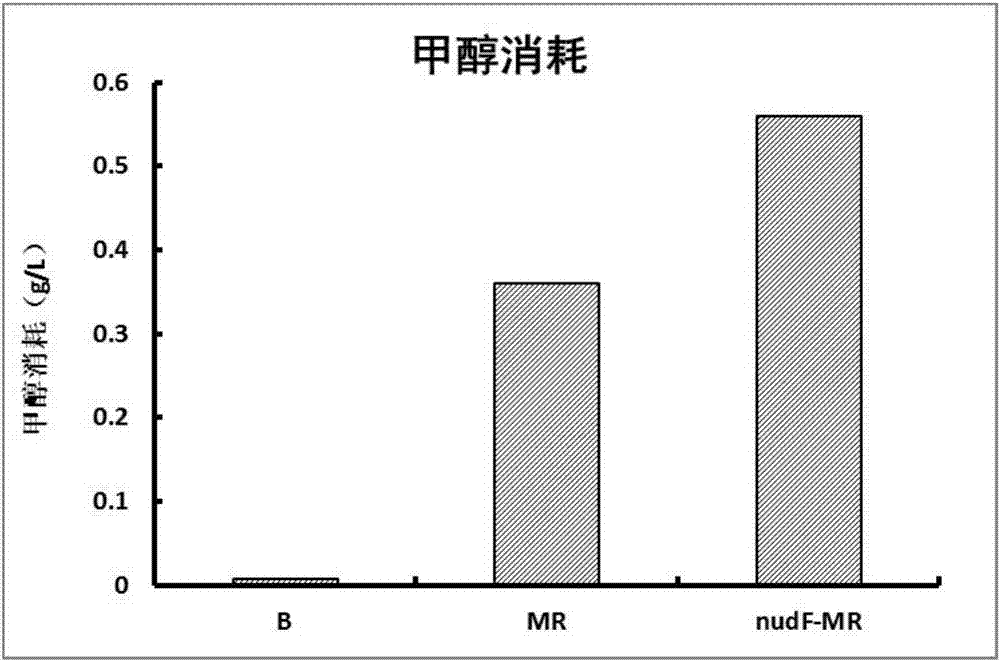
[0028] The invention assembles the methanol metabolism pathway dependent on NAD methanol dehydrogenase and RuMP pathway in Escherichia coli. Mdh2 derived from Bacillus methanolicus MGA3 was selected by comparing methanol dehydrogenase Mdh from different sources with 3-hexose-6-phosphate synthase Hps and 3-hexose-6-phosphate isomerase Phi and Hps-Phi, after the gene was synthesized, the gene was cloned into the plasmid pETDuet, and the plasmid pETDuet-Mdh2 and the plasmid pETDuet-Mdh2-RuMP were constructed. The plasmid was introduced into Escherichia coli BL21(DE3) by conventional methods, and the obtained strains were named M and MR, respectively, and were stored in the form of glycerol tubes, wherein the strain M only overexpressed the gene Mdh2, and was connected to the enzyme cutting site of the vector The dots are NcoI and BamHI; strain MR overexpresses genes Mdh2 and Hps-Phi, and the restriction sit...
Embodiment 2
[0029] Embodiment 2: Enhancing the enzymatic activity of methanol dehydrogenase
[0030] In order to enhance the activity of methanol dehydrogenase Mdh, three schemes have been adopted in this embodiment:
[0031] Scheme 1 is to enhance the enzyme activity of methanol dehydrogenase through directed evolution, but the enzyme activity is not significantly improved;
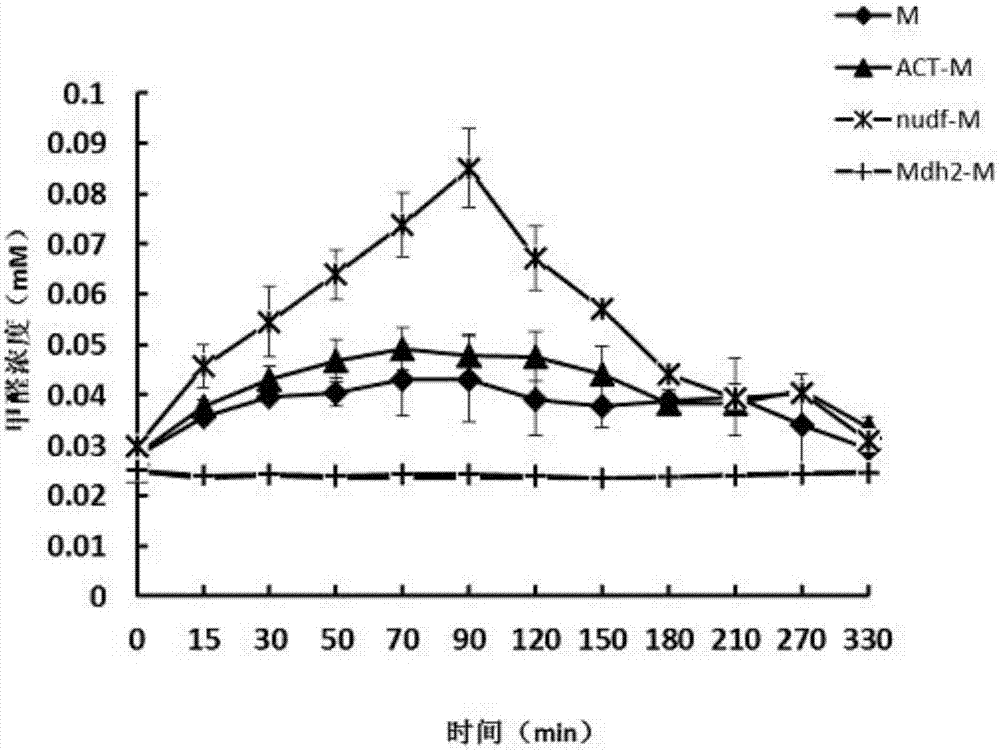
[0032] The second scheme is to enhance the enzymatic activity of methanol dehydrogenase by adding activator protein, in which the activator protein adopts two sources: one is to add the homologous activator protein nudF from E.coliMG1655 to construct the strain BL21(DE3) / pETDuet-nudF -Mdh2 and BL21(DE3) / pETDuet-nudF-Mdh2-RuMP, the strains were named nudF-M and nudF-MR, respectively;
[0033] The second is to add heterologous activating protein ACT from Bacillus.methanolicusPB1 to construct strain BL21(DE3) / pETDuet-ACT-Mdh2, and the strain is named ACT-M;
[0034] Scheme 3 is to replace methanol dehydrogenase Mdh2....
Embodiment 3
[0036] Embodiment 3: the detection of methanol dehydrogenase enzymatic activity
[0037] In the present invention, the enzyme activity of detecting methanol dehydrogenase Mdh adopts the color reaction of Nash reagent (i.e. acetylacetone reagent) and formaldehyde, and detects the activity of methanol dehydrogenase (i.e. the ability of methanol to be oxidized to formaldehyde) by the amount of formaldehyde generated. .
[0038] M9 medium: 17.1g / LNa 2 HPO 4 12H 2 O, 3g / LKH 2 PO 4 , 10g / LNH 4 Cl, 0.5g / LNaCl, trace elements, and 10g / LGlucose.
[0039] Nash reagent: 150g / L ammonium acetate, 3ml / L acetic acid, 2ml / L acetylacetone.
[0040] The strain in the glycerol tube was connected to the shaking tube to rejuvenate the strain, cultivated overnight at 37°C, 200rpm, collected the bacteria, centrifuged at 5000g, 7min, resuspended in M9, transferred to 9ml of M9 medium, added 0.5M methanol to start the reaction, different Time sampling, mixed with Nash reagent 1:1, reacted at 5...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com