Biological genome sequence-based microRNA predicting method
A genome sequence and prediction method technology, applied in the field of microRNA prediction based on biological genome sequence, can solve the problems of high price and time-consuming molecular biology methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
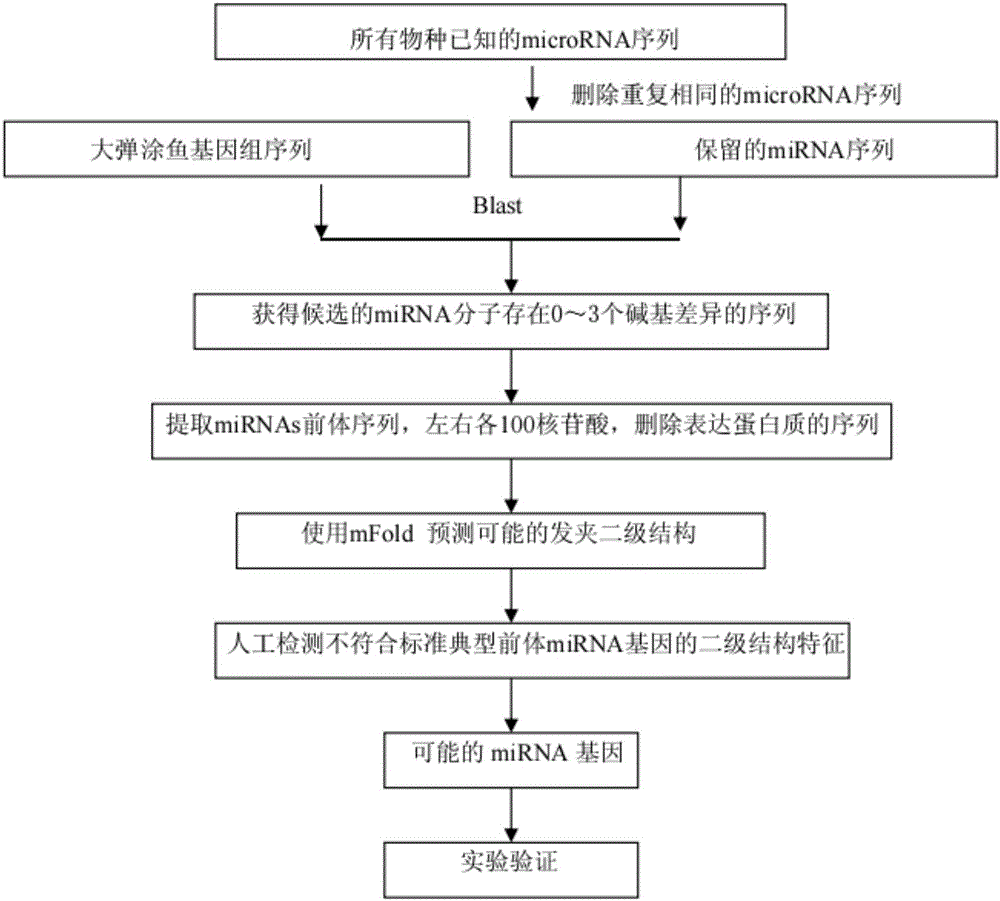
[0021] Such as figure 1 , figure 2 As shown, a kind of microRNA prediction method based on biological genome sequence provided in this embodiment, the specific steps are as follows:
[0022] S1, download the genome sequence and mature miRNAs sequence of biological species from the database in the website;
[0023] S2. Perform blast comparison between the downloaded miRNA sequence and the genome sequence of the species, and search for homologous sequences with 0-3 base differences;
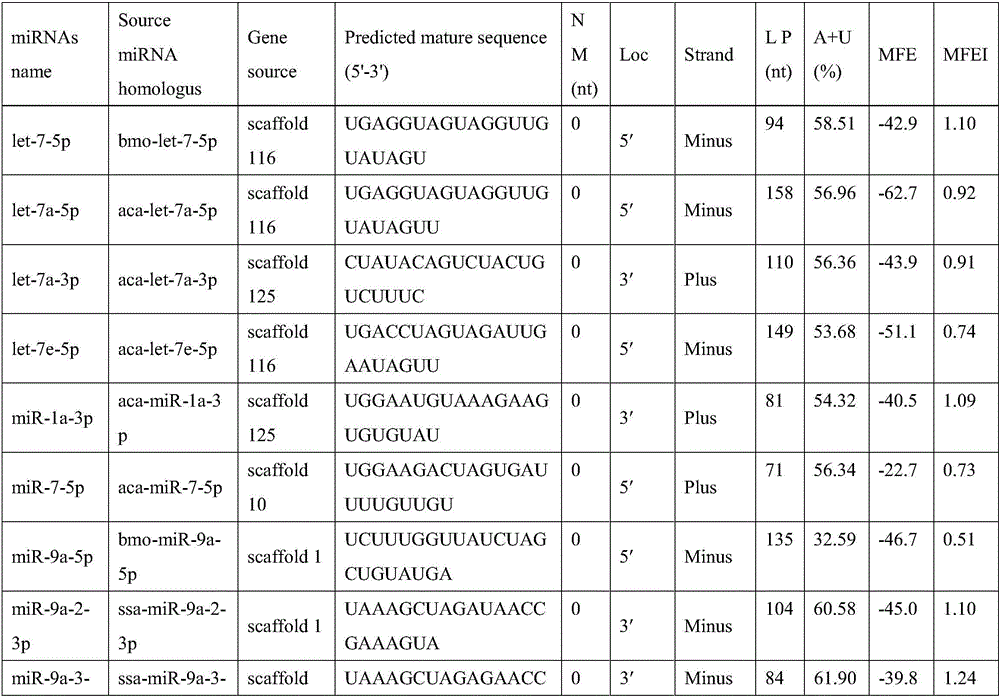
[0024] S3. Extract the microRNA precursor sequence to predict the stem-loop secondary structure, and delete the stem-loop sequence containing the encoded protein;
[0025] S4. Deleting sequences with inappropriate AU content and not conforming to the typical precursor microRNA stem-loop structure;
[0026] S5, through the screening step, obtain the existing microRNA molecules in the large mudskipper;
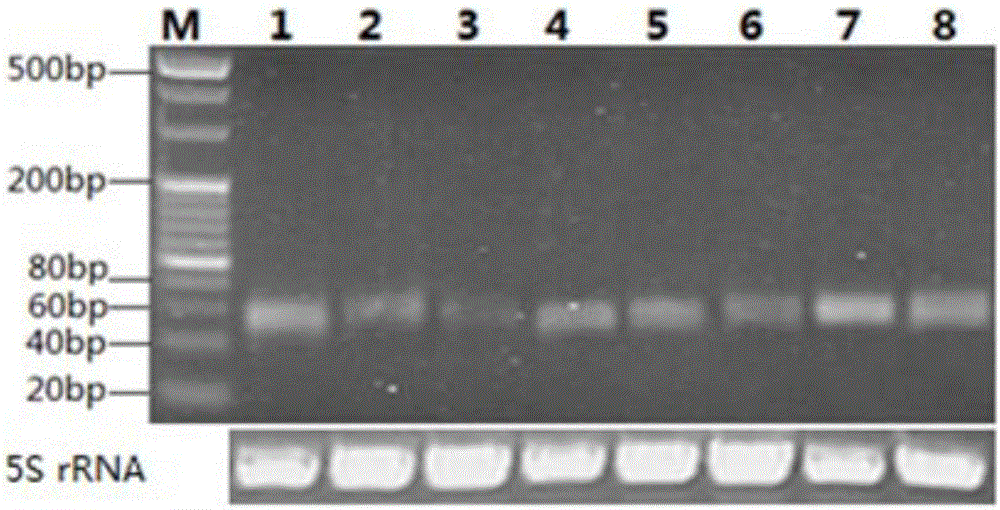
[0027] S6. Using stem-loop RT-PCR experiment to verify the predicted microRNA experimentally...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com