Detection method of specific nucleic acid fragment based on CRISPR-Cas13a
A nucleic acid fragment and detection method technology, applied in the field of biological material detection, can solve problems such as easy contamination of samples, and achieve the effect of improving sensitivity and simplifying operation procedures
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
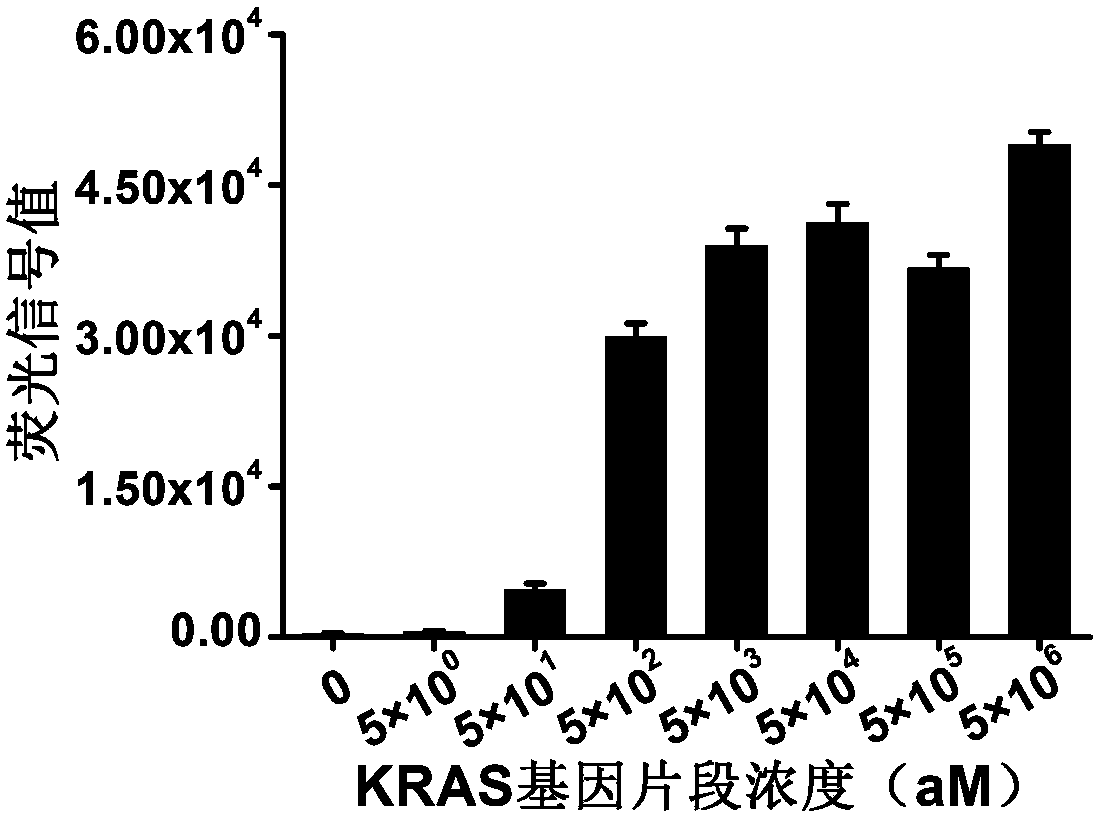
[0054] In this example, the KRAS gene is used to illustrate the detection of the target gene by the detection method (three-in-one) of the present invention, and to test the sensitivity of the method.
[0055] First, by annealing after primer synthesis, a fragment of the KRAS gene was synthesized as the target double-stranded DNA for detection.
[0056] Secondly, RPA primers were synthesized, the upstream primer was 5'-taatacgactcactataggctgctgaaaatgactgaatataaactt-3' (the 19 nucleotides at the 5' end were T7 promoter), and the downstream primer was 5'-ggatcatattcgtccacaaaatgattct-3'.
[0057] Again, by in vitro transcription, a crRNA targeting the KRAS gene fragment was synthesized with the sequence 5'-GACCACCCCAAAAATGAAGGGGACTAAAACccaccagctccaactaccaagtttat-3', where the uppercase letters are the anchor sequence and the lowercase letters are the guide sequence, which recognizes the 5'- ataaacttgtggtagttggagctggtgg-3'.
[0058] Finally, construct a 50 μL reaction system:
...
Embodiment 2
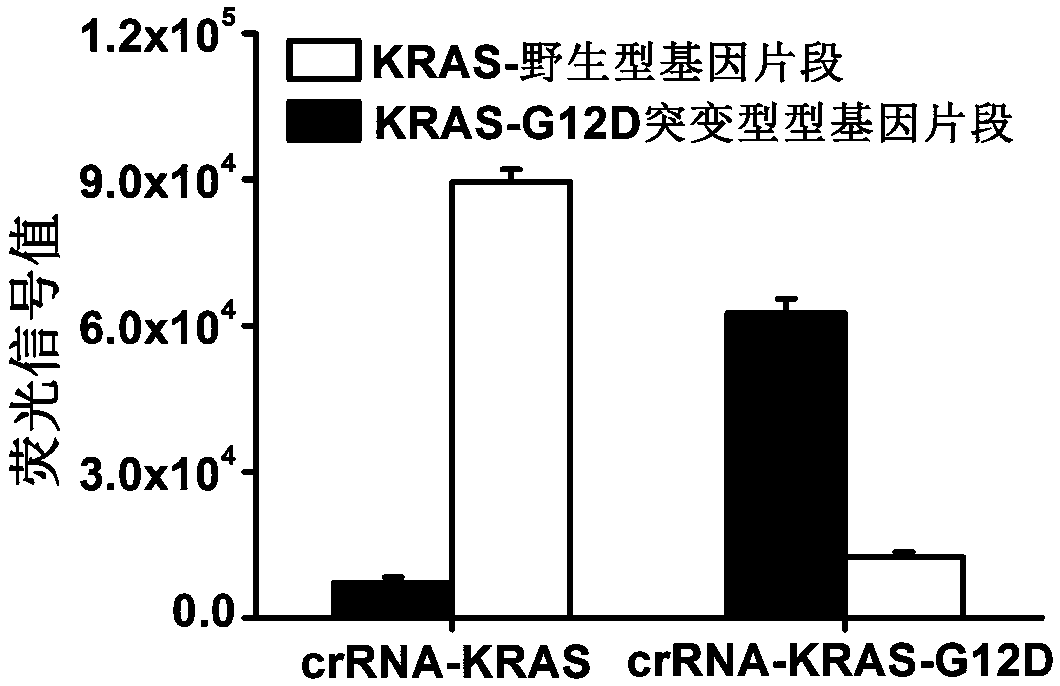
[0077] In this example, the KRAS gene and the KRAS-G12D mutant gene are used as the target genes to illustrate the detection method (three-in-one) of the present invention for the detection of target genes, and to test the specificity of the method.
[0078]首先,通过引物合成后退火的方法,分别合成了KRAS基因片段以及KRAS-G12D突变片段作为检测的目的双链DNA,序列分别为5'-ctgctgaaaatgactgaatataaacttgtggtagttggagctggtggcgtaggcaagagtgccttgacgatacagctaattcagaatcattttgtggacgaatatgatcc-3'和5'-ctgctgaaaatgactgaatataaacttgtggtagttggagctgAtggcgtaggcaagagtgccttgacgatacagctaattcagaatcattttgtggacgaatatgatcc-3',其中 Capital A is the mutant base.
[0079] Secondly, RPA primers were synthesized, the upstream primer was 5'-taatacgactcactataggctgctgaaaatgactgaatataaactt-3' (the 19 nucleotides at the 5' end were T7 promoter), and the downstream primer was 5'-ggatcatattcgtccacaaaatgattct-3'.
[0080] Thirdly, since the Cas13a protein can generally tolerate one base mismatch, we synthesized the crRNA-KRAS targeting the KRAS wild-type gene fragment by...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com