Anti-CD19 antibody and preparation method and use thereof
A CDR-H1, CDR-L1 technology, applied in the field of anti-CD19 antibody and its preparation, can solve the problems of poor persistence of CART cells, decreased affinity, and inability of CART cells to clear plasma cells
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0094] Construction of CDR1, CDR2, and CDR3 mutation libraries of heavy chain (H) and light chain (L) of FMC63 scFv and C2146 scFv:
[0095] The selection of FMC63 and C2146 heavy chain and light chain CDR regions is based on the amino acid counting method of the variable region (the template is the monoclonal antibody FMC63, VH: Y14283.1, VL: Y14284.1; the C2146 single-chain antibody sequence is selected from US20140271635), Kabat counts (Kabat number scheme.Bioinf.org.uk) are listed below:
[0096] Table 1
[0097]
[0098]
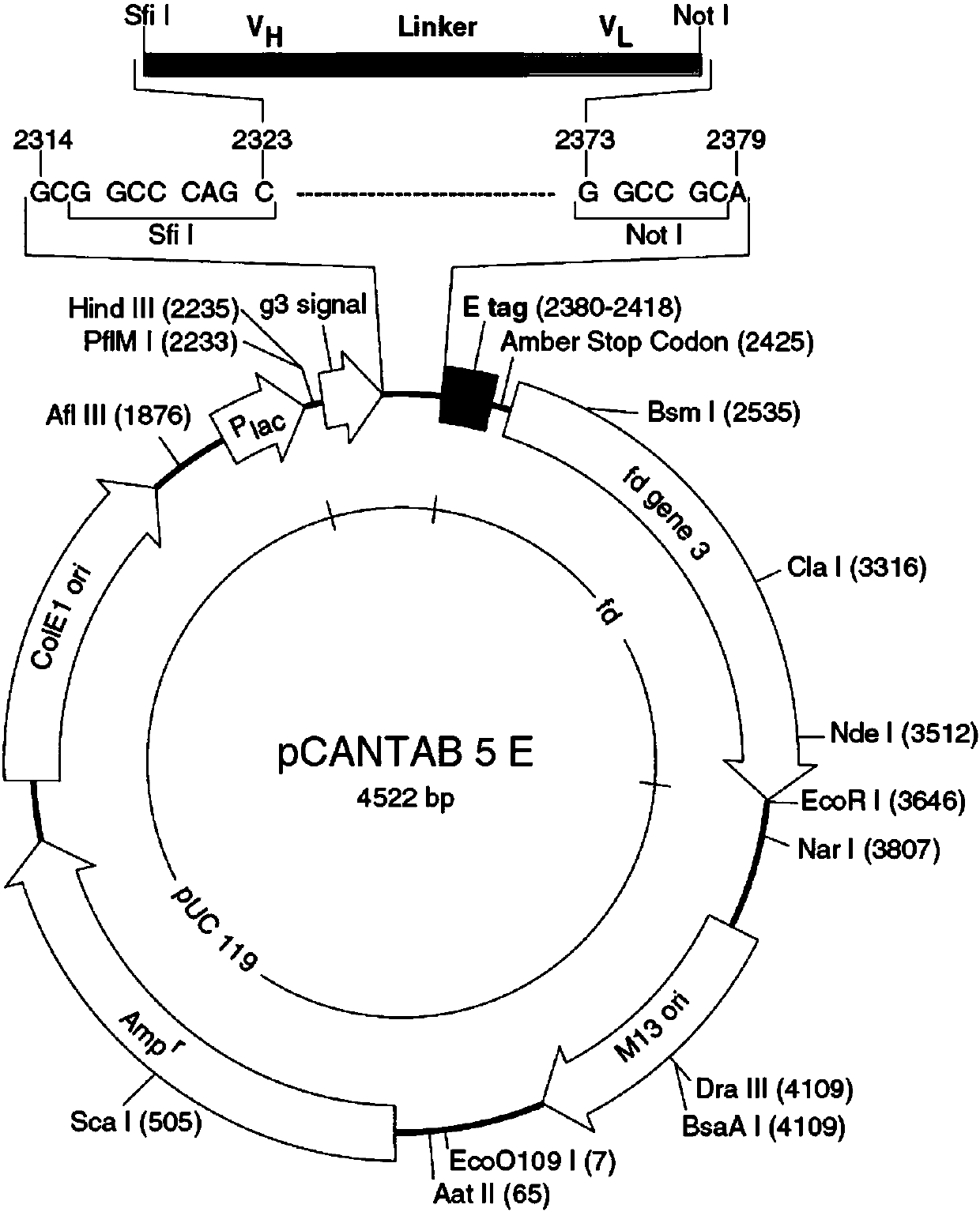
[0099] Constructed with pCAN-FMC63 scFv plasmid and pCAN-C2146 scFv (the template sequence is inserted into the multiple cloning site of pCANTAB 5E plasmid (purchased from GE), the FMC63 template sequence is: VH: Y14283.1, VL: Y14284.1, C2146 template See US20140271635 for the sequence) as a template, and introduce mutations by PCR with random primers. The primers for the FMC63 mutation library and the C2146 mutation library primers are shown in ...
Embodiment 2
[0107] Panning of phage antibody library:
[0108] Add 20nM CD19-his-biotin antigen and FMC63 scFv or C2146 scFv phage antibody library and incubate at room temperature for 2h, then transfer the mixture to streptavidin magnetic beads (Dynabeads M-280Streptavidin) and incubate for 15min at room temperature. Unbound phages were washed away with PBST-PBS, and trypsin was added for 30 min at 37°C to elute bound phages. Infect 4ml of TG1 cells in the logarithmic phase with the phages eluted by trypsin digestion, let stand at 37°C for 30min, and take part of the bacterial liquid to serially dilute (1 / 10, 1 / 100, 1 / 1000) for plate counting, and the remaining bacteria The solution was all coated on 2xYT(GA) plates for the next round of packaging. The packaged phage can be used for the next round of panning. A total of 4 rounds of panning and enrichment were carried out. Each round of panning was 10 times diluted and the antigen concentration was gradually reduced, and the number of PB...
Embodiment 3
[0110] Screening and identification of high-affinity scFv:
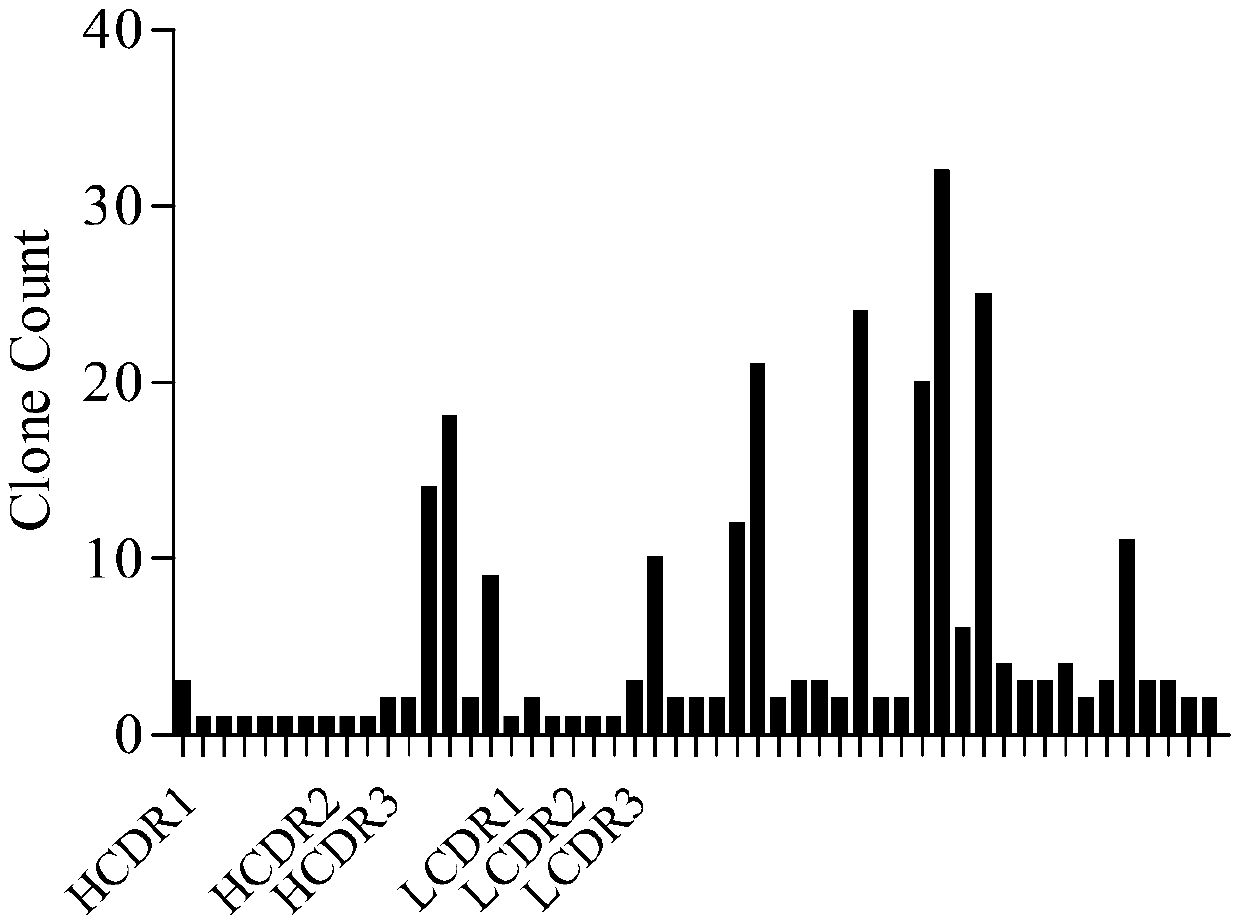
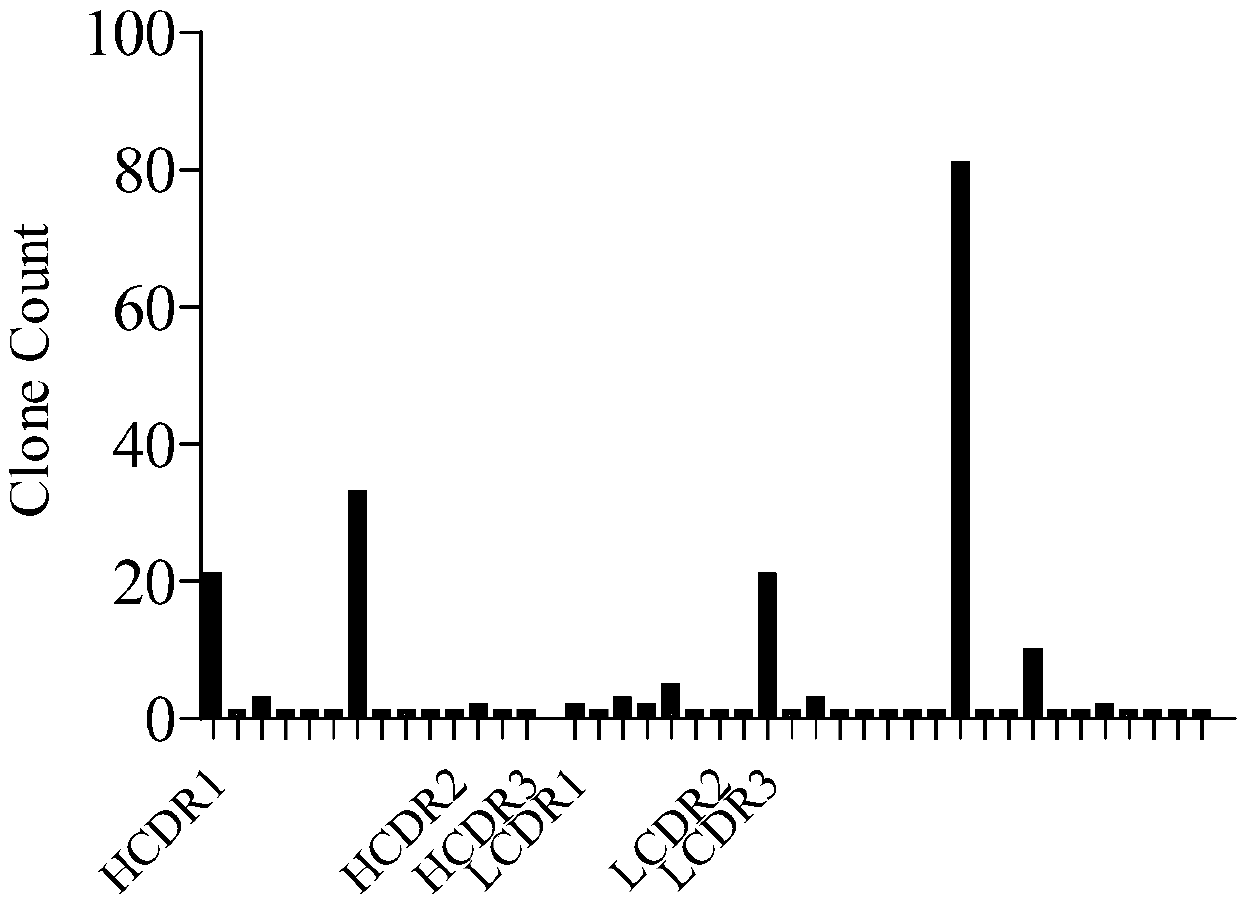
[0111]After four rounds of panning, single clones were randomly selected, and the supernatant was taken for ELISA screening after IPTG induction: 1ug / ml Avidin was coated overnight on a 96-well plate, washed twice with PBST, and 100ul 1ug / ml CD19-biotin was added Avidin-coated 96-well plates were incubated for 1 hour, washed twice with PBST, incubated with supernatant after IPTG induction for 1 hour, washed 4 times with PBST, added 100ul diluted anti-Myc antibody at 1:5000, incubated for 30 minutes, washed with PBST After 4 times, add TMB substrate to react for 10 minutes, add 1M sulfuric acid solution to terminate the reaction, and read OD450nm light absorption. Pick the clones whose positive signal (OD450nm light absorption) is at least 2 times greater than the negative signal and send them for sequencing, analyze the sequencing results, and extract the clones corresponding to the more enriched CDR regions. The re...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com