Methods of identifying and validating affinity reagents
A specific, cell-based technology, applied in the field of affinity reagents, can solve the problems of high cost and low throughput of display technology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
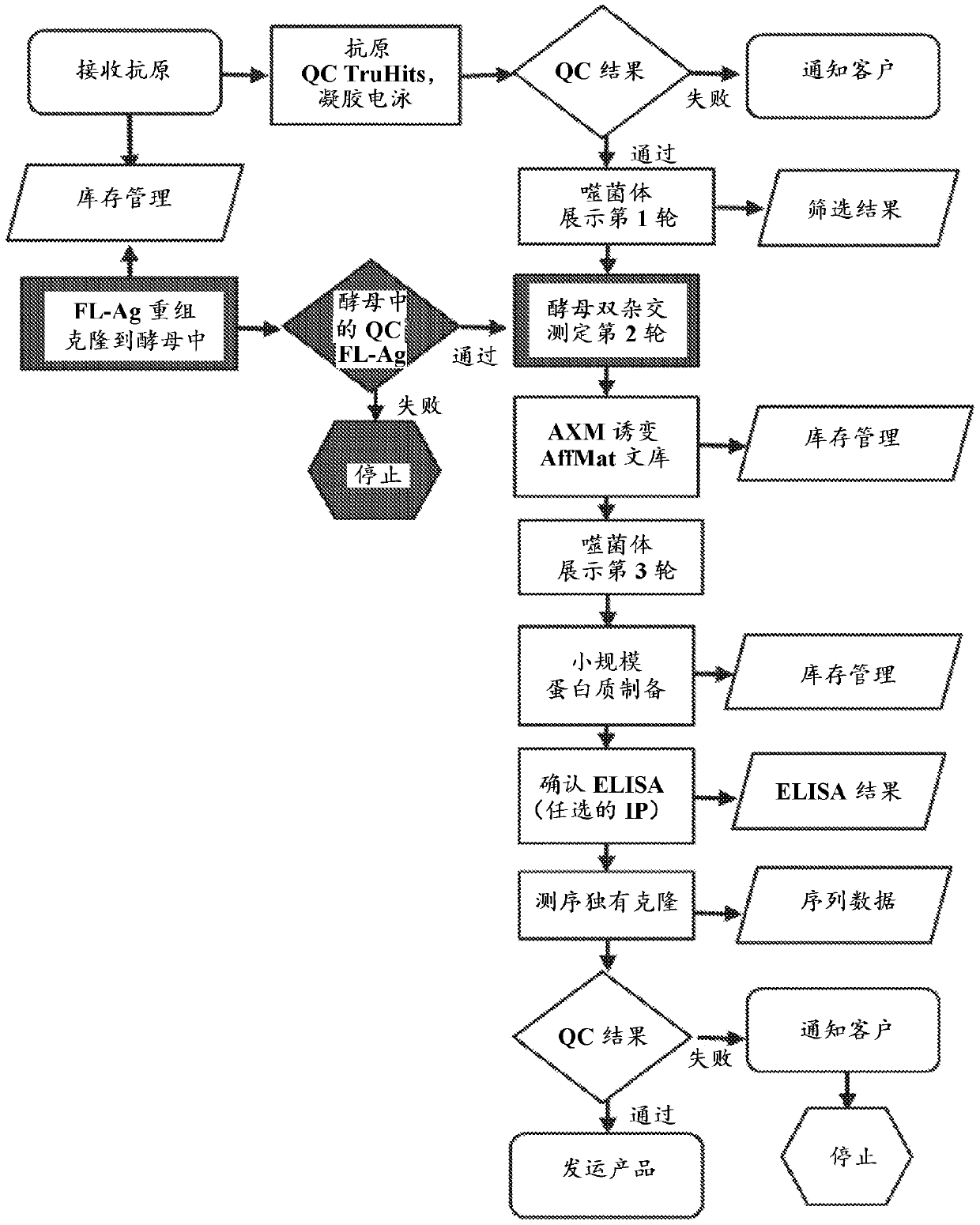
[0107] Example 1. Materials and methods
[0108] Bacterial Strains and Vectors
[0109] Escherichia coli ( E. coli ) strain TG1[F'( tra D36, pro AB+ lac I q , lac ZΔM15), sup E, thi -1, Δ( lac-pro AB), Δ( mcr B- hsd SM) 5, (rK - mK - )] were purchased from Lucigen (Middleton, WI) and used to encode Eco The pAX1492 plasmid of the 29kI RM operon was transformed into TG1 to develop E. coli ( E. coli ) strain AXE688. Escherichia coli ( E. coli ) strain CJ236 (FΔ (Hin DIII )::cat( Tra + , Pil + ,Cam R ) / ung-1, relA1, dut-1, thi- 1, spoT1, mcrA ) were purchased from New England BioLabs (NEB; Waverly, MA). Electrocompetent and chemically competent strains will be prepared according to the NEB protocol. The template plasmid used for AXM mutagenesis was a derivative of the phagemid pCH103, and the same scFv template was genetically fused to the coat protein gp3 of phage M13. Each of the six CDRs of this template scFv will be modified to include...
Embodiment 2
[0124] Example 2. Overview of antibody framework and library design
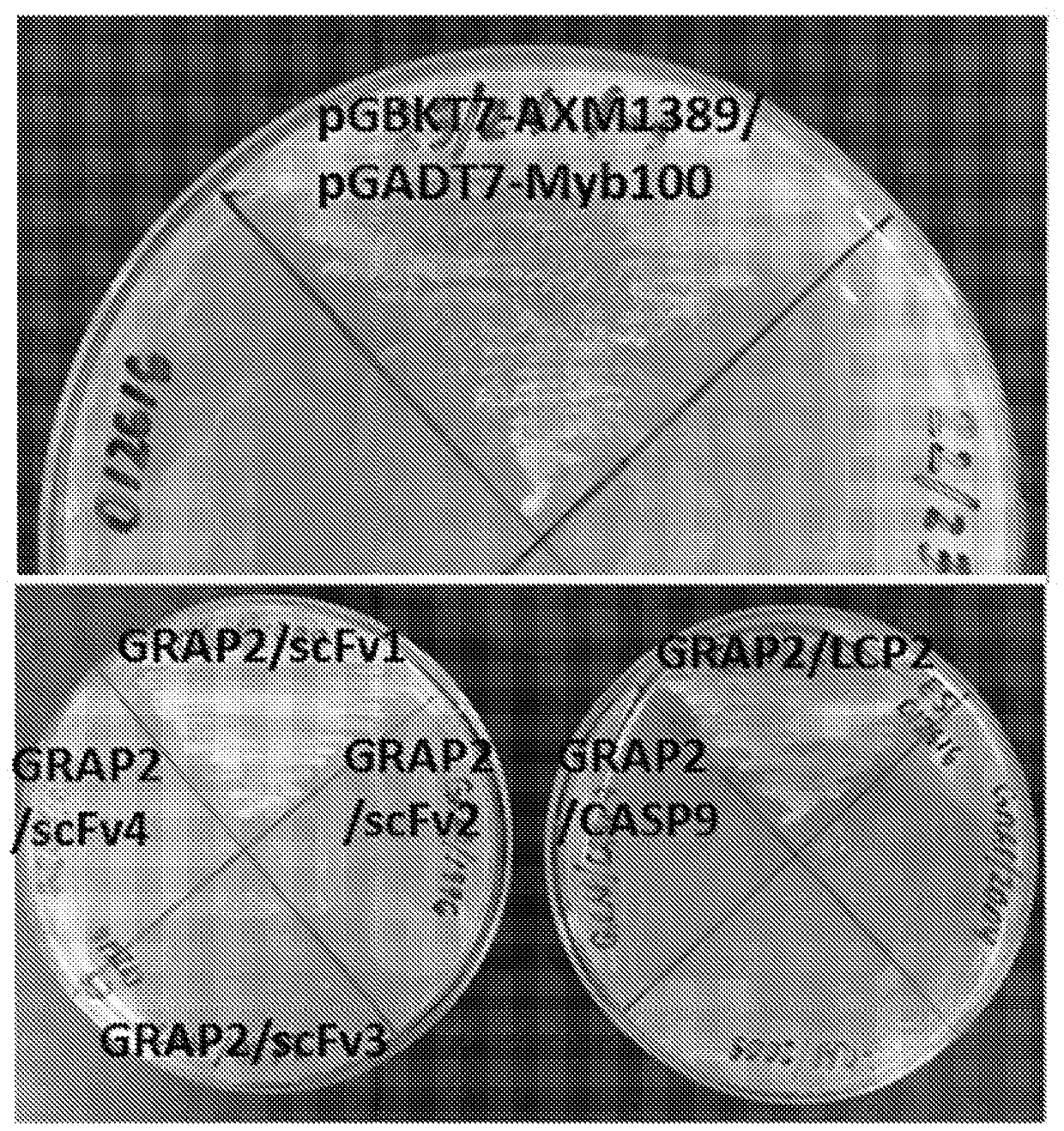
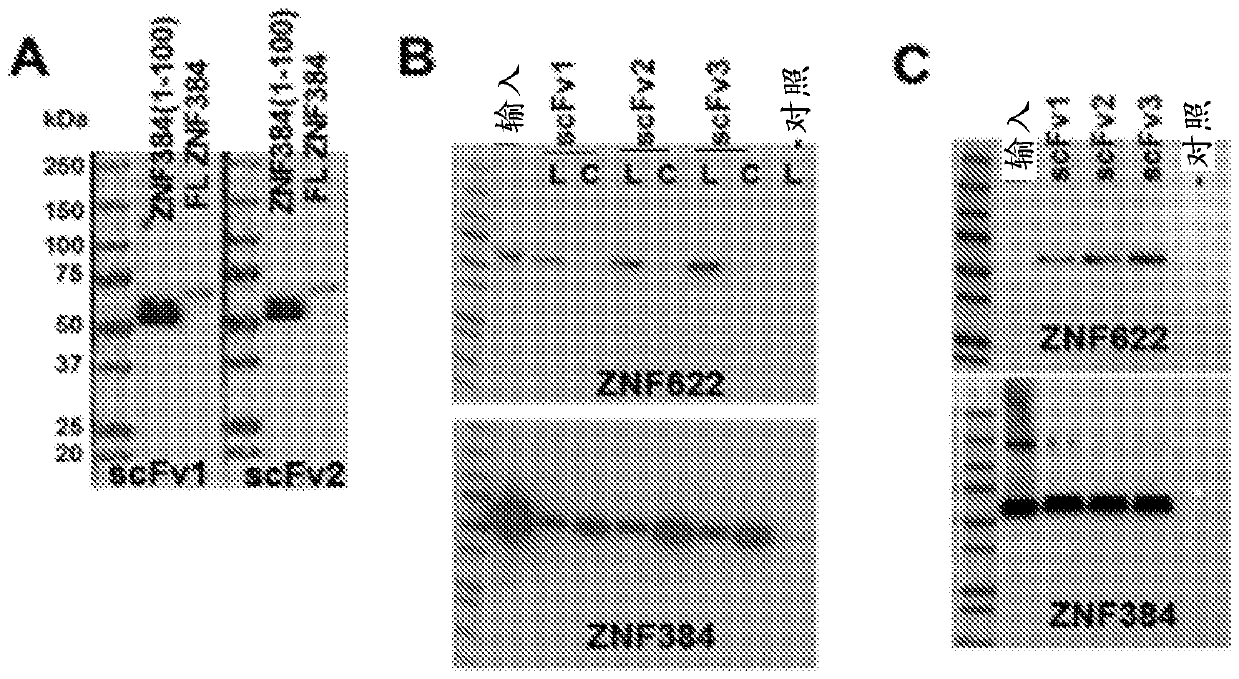
[0125] We have identified Ab libraries based on constant framework scFvs that encode the NNK codon (see e.g. Benhar, Expert Opin Biol Ther. 7(5):763-79, 2007; Chan et al., Int Immunol. 26(12): 649-657, 2014; Miersch et al., Methods.57(4):486-98, 2012; Mondon et al., Front Biosci.13:1117-29, 2008; Tohidkia et al., J Drug Target.20(3) :195-208, 2012), with coding positions at 18 different positions within the six complementarity-determining regions (CDRs) (Mandrup et al., PLoS One. 8(10):e76834, 2013). The B2A framework of the library identified and used in these experiments was based on its ability to function in phage display (ɸD) (when fused to the M13 gp3 protein) and yeast two-hybrid (Y2H) (when fused to the activation domain) Make a selection. It is worth noting that although the platform of the present invention for generating rAbs is currently used for single chain variable fragments (scFv), this pla...
Embodiment 3
[0131] Example 3.pCH103 cloning vector system can eliminate the pair used in ɸD, yeast two-hybrid, and Escherichia coli (E. coli) The need for subcloned proteins tested in protein expression
[0132] Techniques for Y2H and ɸD can be adapted to develop selective and scalable systems for obtaining, characterizing and affinity-matured antibodies in a very high-throughput and cost-effective manner. A dual-function vector system (pCH103) has been constructed, which has both Y2H and Escherichia coli ( E. coli ) the ability to function in the ɸD approach ( figure 2 ). in Escherichia coli ( E. coli ), protein expression depends on E. coli ( E. coli )genotype. The main features of the pCH103 vector system and its beneficial effects are described below.
[0133] pCH103 vector design
[0134] like figure 2 As shown in A, an amber stop codon was inserted between the protein of interest (scFv in this case) and gp3. Therefore, in non-inhibitory E. coli ( E. coli ) middle , ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com