Multi-PCR (Polymerase Chain Reaction) primer design method based on Primer 3
A primer design, multiplexing technology, applied in biochemical equipment and methods, recombinant DNA technology, microbial assay/inspection, etc. Problems such as high-frequency polymorphism sites of single nucleotides cannot be avoided to achieve the effect of reducing non-specific amplification and high specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
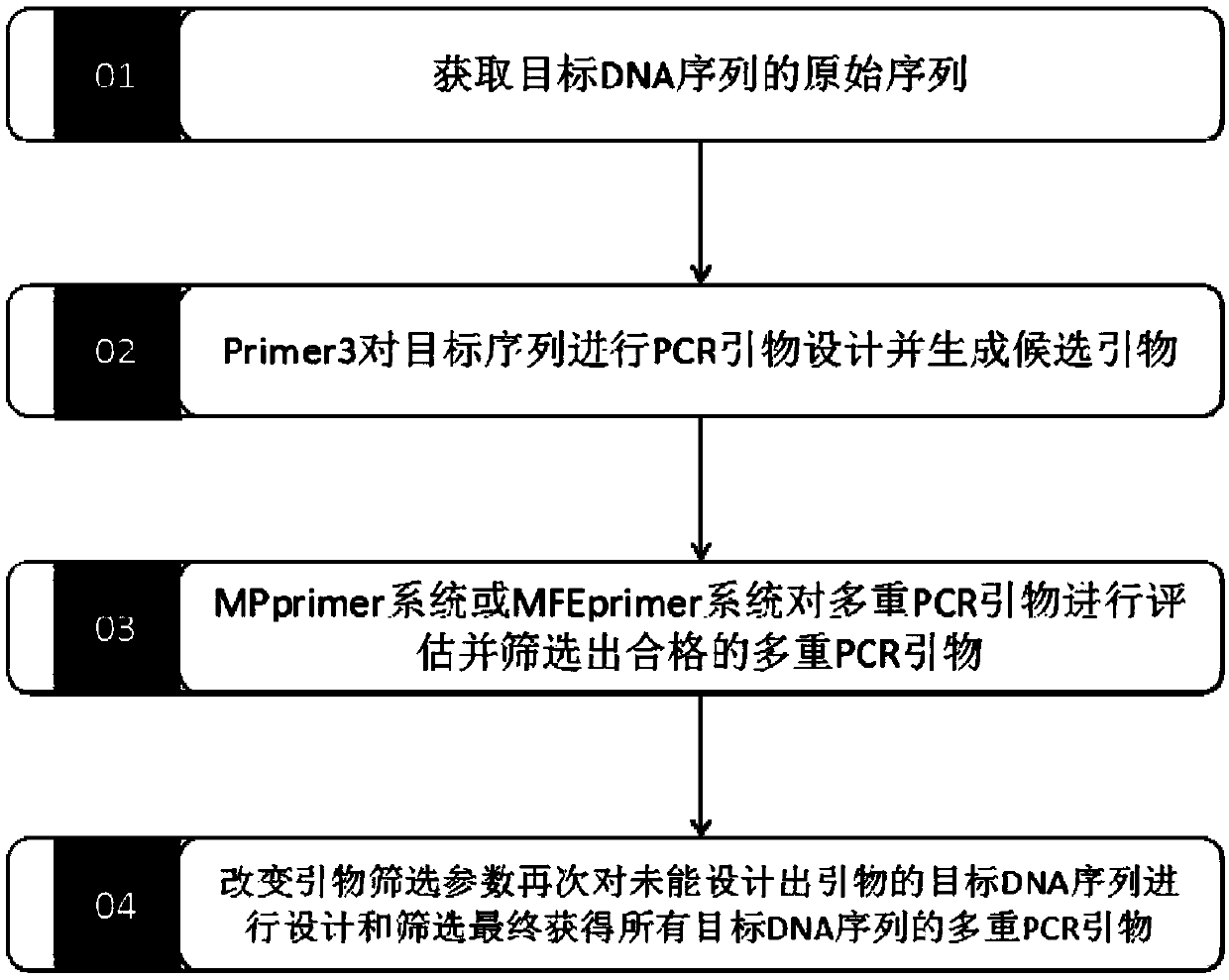
[0056] This embodiment provides a method for designing multiplex PCR primers based on Primer3, such as figure 1 shown, which includes:
[0057] S1: Obtain the original sequence of the target DNA sequence;
[0058] S2: Primer3 designs PCR primers for the target sequence and generates candidate primers;
[0059] S3: Evaluate multiplex PCR primers with PE model or SE model, and screen out qualified multiplex PCR primers;
[0060] S4: Change the primer screening parameters, design and screen again the target DNA sequences for which no primers have been designed, and finally obtain multiple PCR primers for all target DNA sequences.
[0061] This example provides a multiplex PCR primer design method based on Primer3, which solves the problems of amplification failure caused by mismatching between primers and DNA templates, non-specific amplification, primer dimers and hairpin structures, etc., the system Consistently evaluate the specificity of the designed primers to ensure the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com