Amplification primer pair for lactobacillus sequencing, lactobacillus species identification method and application
A technology for amplifying primers and lactic acid bacteria, applied in the field of genetic engineering, can solve problems such as research, unsatisfactory, unfavorable lactic acid bacteria community structure, etc., and achieve the effects of good diversity, good specificity and practicability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0035] Use the above-mentioned amplification primers of the present invention to carry out PCR amplification to the lactic acid bacteria of 7 genera and 14 species, specifically comprising the following steps:
[0036] The DNA of 14 strains of 7 genera of common lactic acid bacteria was used as the positive template, and the sterilized and filtered ultrapure water was used as the negative template, and Lab-6F and Lab-6R were used as primers for PCR amplification to obtain PCR amplification products;
[0037] The nucleotide sequences of the primer pairs are as follows:
[0038] Lab-6F: 5'-GCTCAGGAYGAACGCYGG-3', where Y represents C;
[0039] Lab-6R: 5'-CACCGCTACACATGRADTTC-3', where R represents A and D represents T.
[0040] Among them, the 14 bacterial strains are Enterococcus asini (DSM-11492), Enterococcus faecium (ATCC 19434T), Enterococcus Italicus (DSM-15952), Lactobacillus Casei (ATCC-334), Lactobacillus Salivarius (DSM-20555), Lactobacillus Helveticus (1.2278) , Weis...
Embodiment 2
[0047] Compare lactic acid bacteria amplification primer Lab-6 (Lab-6F and Lab-6R) of the present invention and bacterial universal primer 27f / 1492r to the amplification result of lactic acid bacteria in the stool sample, specifically comprise the following steps:
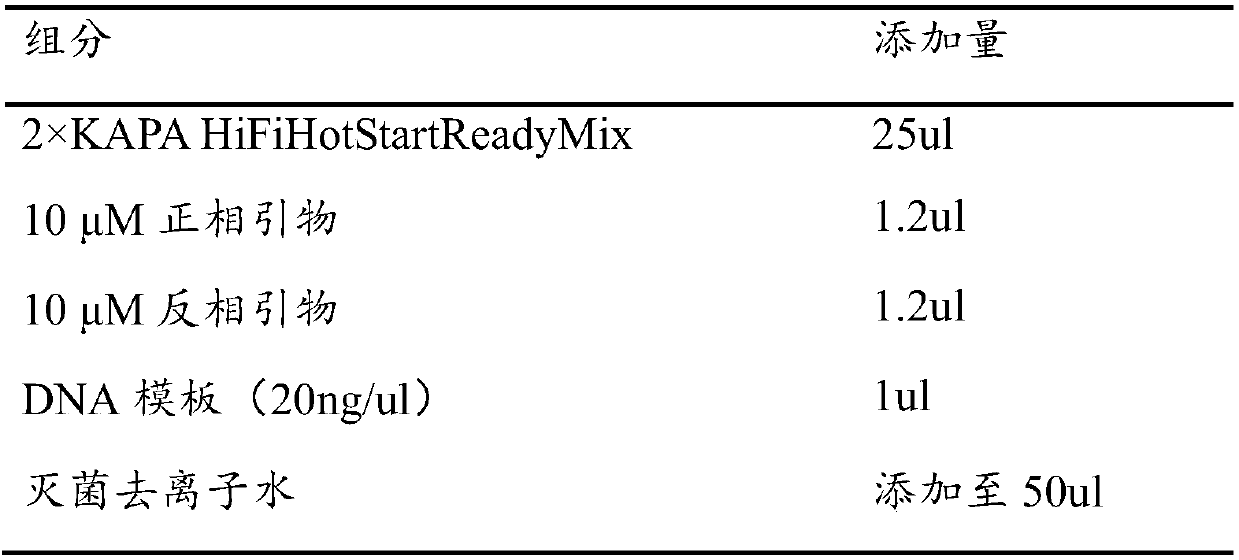
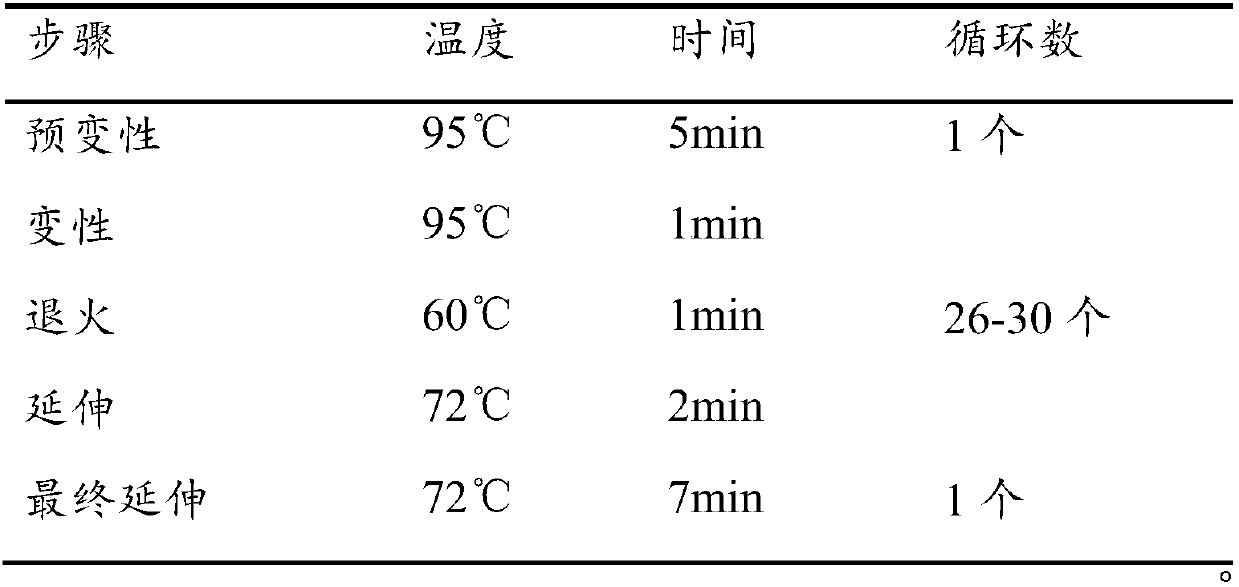
[0048] (1): Amplify the stool DNA dilution with the bacterial universal primer (27f / 1492r) and the amplification primer pair provided in S1 (the nucleotide sequence of the primer pair is the same as in Example 1), and perform PCR amplification System and PCR amplification reaction procedure are all identical with embodiment 1, obtain PCR amplification product;
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com