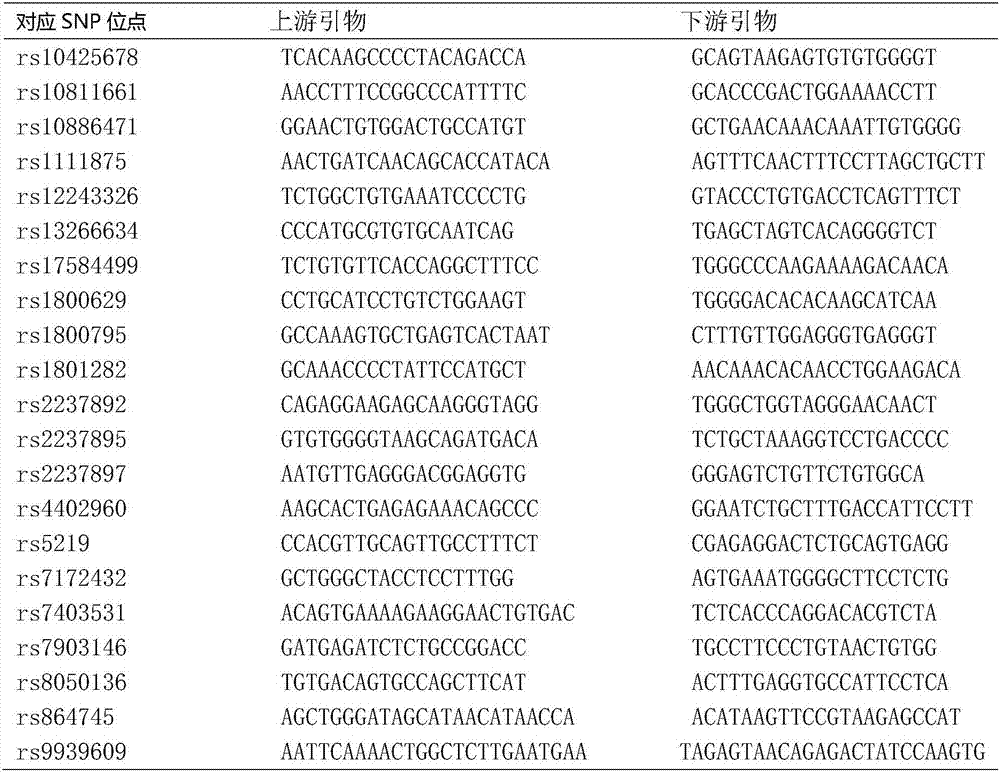
Mutation site combination and primer for detecting susceptible genes of type 2 diabetes as well as application of combination and primer
A type 2 diabetes and mutation site technology, applied in the field of molecular biology, can solve the problems of warning and guidance, high cost, and low accuracy for potential patients who cannot be diabetic
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0033] This example is used to illustrate the method of using the primer combination described in the present invention to detect the mutation site described in the present invention, and the specific steps are as follows:
[0034] 1. Peripheral blood extraction
[0035] Whole blood was frozen in EDTA anticoagulant tubes at -80°C, and DNA was extracted using the QIAAmpgDNABlood Minikit.
[0036] 1) Take 200 μL blood sample with added anticoagulant to a 1.5 mL centrifuge tube, then add 40 μL proteinase K solution,
[0037] Then add 200μL AL Buffer, and immediately vortex and oscillate thoroughly for 15 seconds;
[0038] 2) Incubate at 56°C for 30 minutes;
[0039] 3) Take out the cracked specimen from the oven and shake it shortly;
[0040] 4) Add 200 μL of absolute ethanol, immediately moisten and vortex to fully mix, and shake briefly;
[0041] 5) Add the mixture of the previous step (including possible precipitation) into an adsorption column (the adsorption column is pl...
Embodiment 2
[0094] In this example, using the method described in Example 1, 1054 diabetic patients were tested for genes, and 1028 of them were identified as type 2 diabetic patients, with a detection accuracy rate of 97.5%. Further, for 512 Asian healthy persons data from the 1000G (Thousand Genomes Project) public database, the genotypes of the 21 mutation sites described in the present invention were analyzed, and the formula in Example 1 was used to calculate according to the analysis results, and judged 451 were non-type 2 diabetes patients, and the detection accuracy rate was 88.1%. The comprehensive detection accuracy rate is 92.8%,
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com