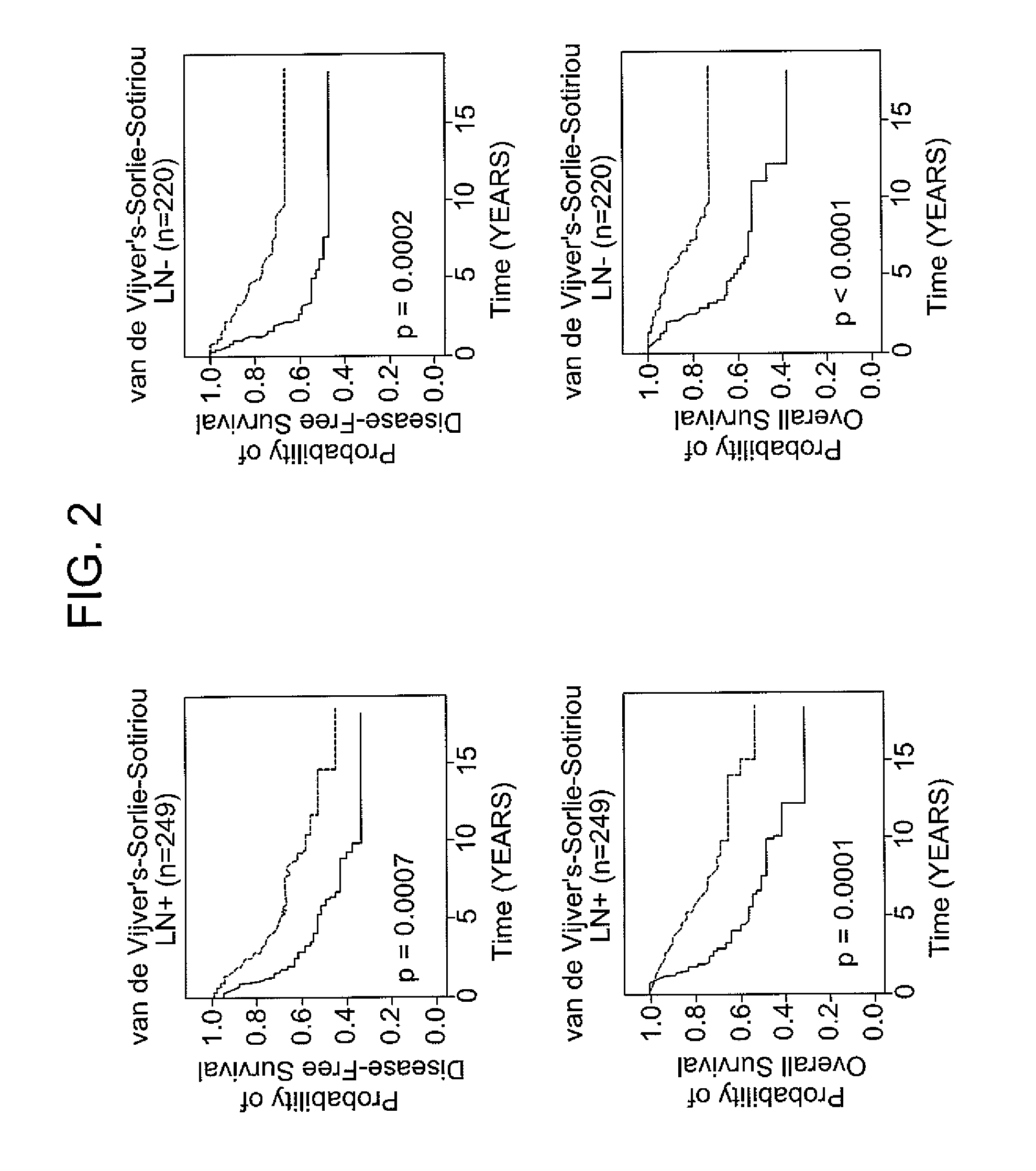
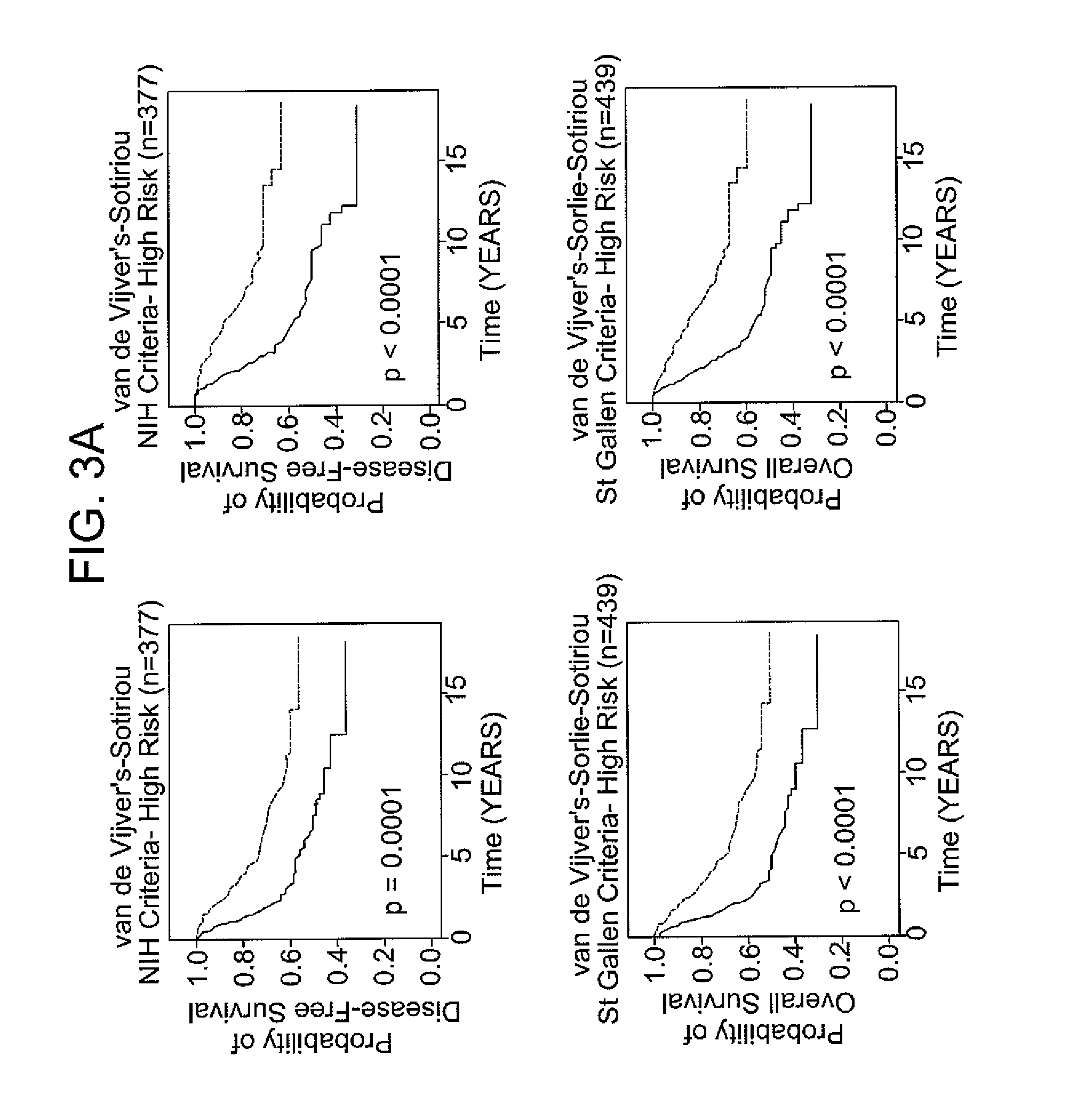
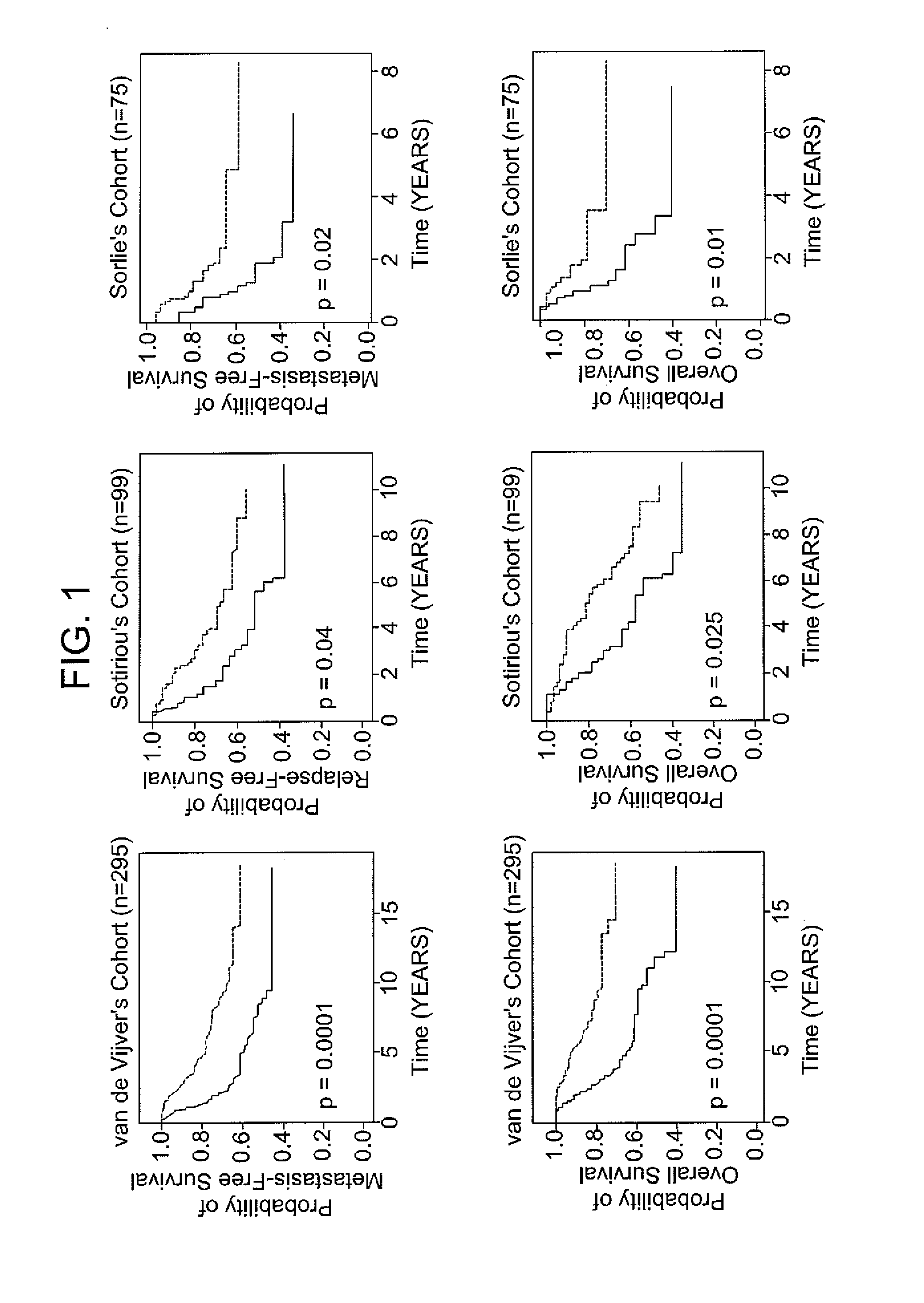
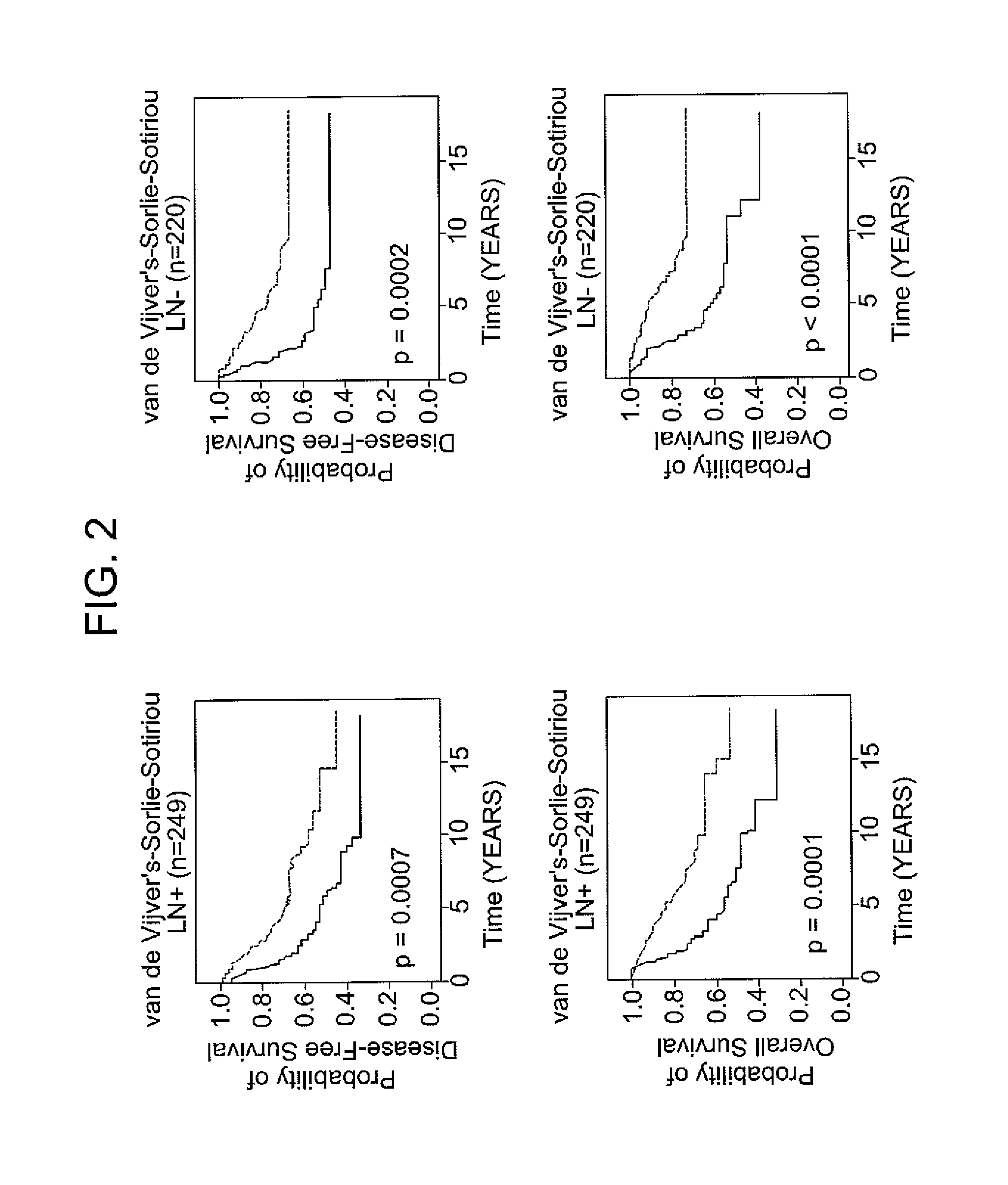
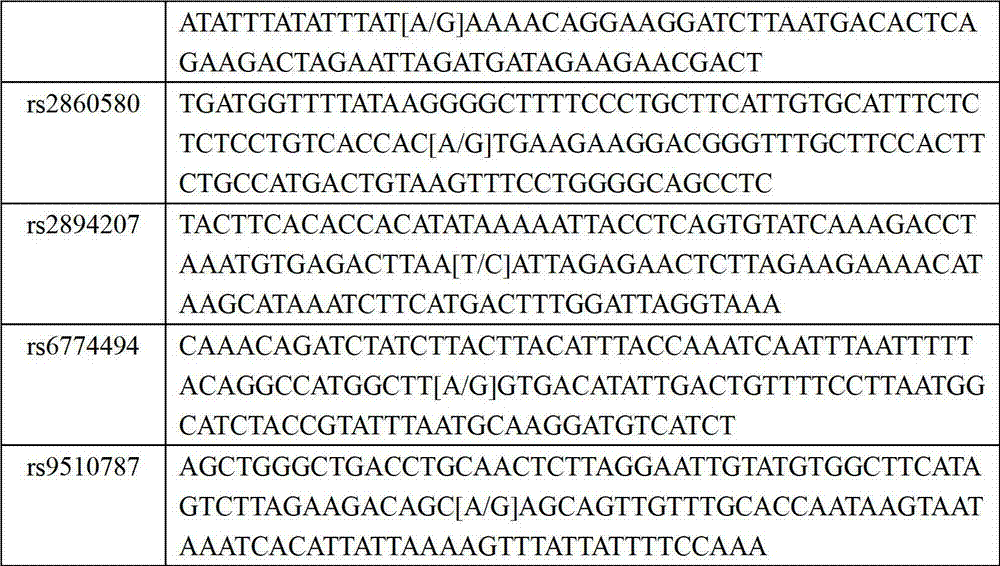Patents
Literature
44 results about "CDKN2A" patented technology
Efficacy Topic
Property
Owner
Technical Advancement
Application Domain
Technology Topic
Technology Field Word
Patent Country/Region
Patent Type
Patent Status
Application Year
Inventor
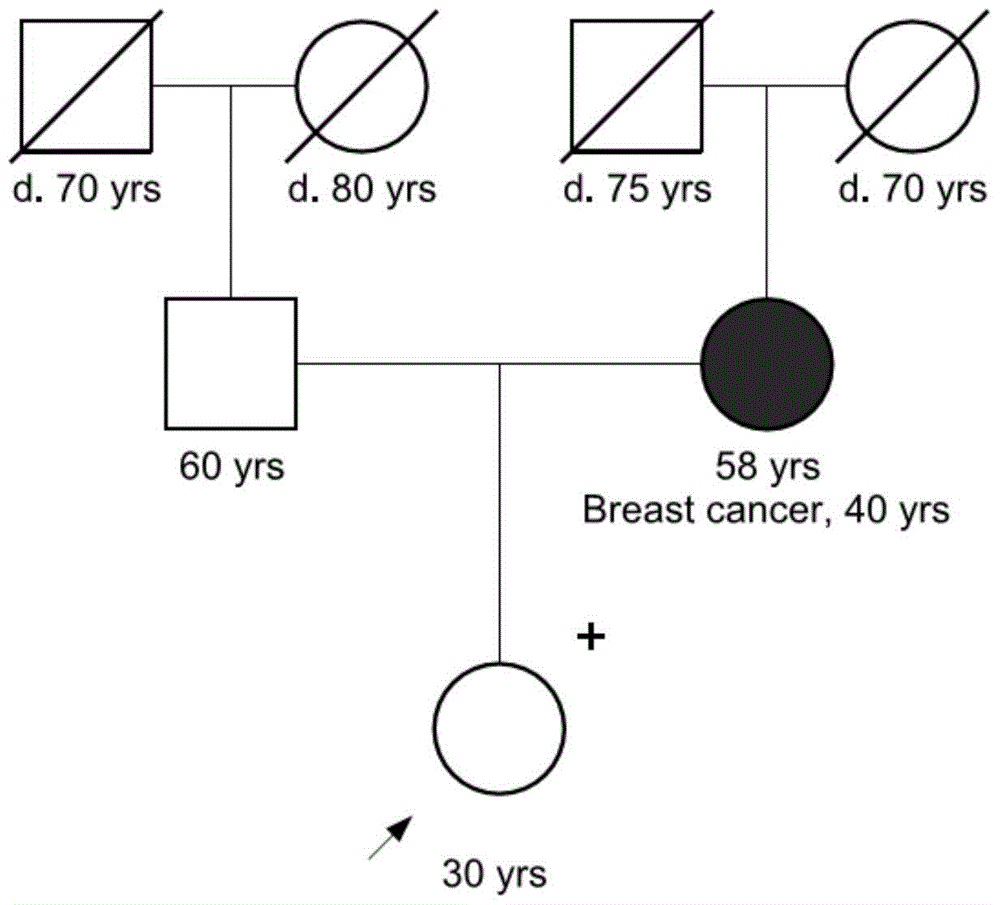
CDKN2A, also known as cyclin-dependent kinase Inhibitor 2A, is a gene which in humans is located at chromosome 9, band p21.3. It is ubiquitously expressed in many tissues and cell types. The gene codes for two proteins, including the INK4 family member p16 (or p16INK4a) and p14arf. Both act as tumor suppressors by regulating the cell cycle. p16 inhibits cyclin dependent kinases 4 and 6 (CDK4 and CDK6) and thereby activates the retinoblastoma (Rb) family of proteins, which block traversal from G1 to S-phase. p14ARF (known as p19ARF in the mouse) activates the p53 tumor suppressor. Somatic mutations of CDKN2A are common in the majority of human cancers, with estimates that CDKN2A is the second most commonly inactivated gene in cancer after p53. Germline mutations of CDKN2A are associated with familial melanoma, glioblastoma and pancreatic cancer. The CDKN2A gene also contains one of 27 SNPs associated with increased risk of coronary artery disease.
Tumor susceptibility 62 genes and application thereof
InactiveCN105986031AImprove the detection rateEasy to identifyHealth-index calculationMicrobiological testing/measurementBAP1MAP2K4
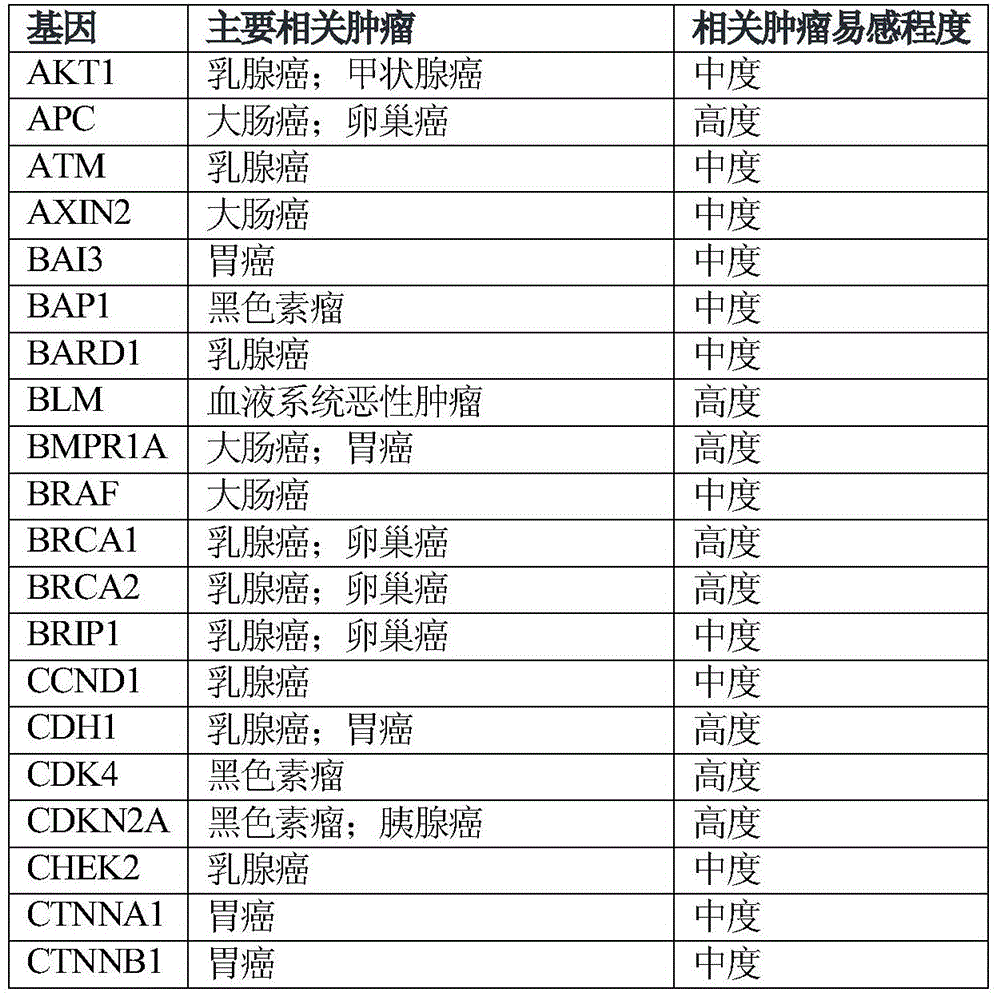
The invention relates to tumor susceptibility 62 genes and application thereof. The tumor susceptibility 62 genes comprise PTEN, STK11, CDH1, TP53, BRCA1, BRCA2, PALB2, CHEK2, ATM, BRIP1, NBN, RAD51C, MLH1, MSH2, MSH6, PMS2, BARD1, RAD51D, MRE11A, MUTYH, PMS1, RAD50, XRCC2, AKT1, PIK3CA, FANCC, RECQL, CCND1, ERBB2, ESR1, GATA3, FGFR1, MAP2K4, MAP3K1, BAI3, CTNNB1, BRAF, KRAS, CTNNA1, EPCAM, APC, BLM, SMAD4, BMPR1A, POLD1, POLE, AXIN2, MEN1, KIT, EGFR, EZH2, PRF1, CDKN2A, CDK4, BAP1, RB1, ERCC2, VHL, MET, FH, FLCN and RET. The detection of the genes can be used for evaluating tumor susceptibility.
Owner:BEIJING CANCER HOSPITAL PEKING UNIV CANCER HOSPITAL
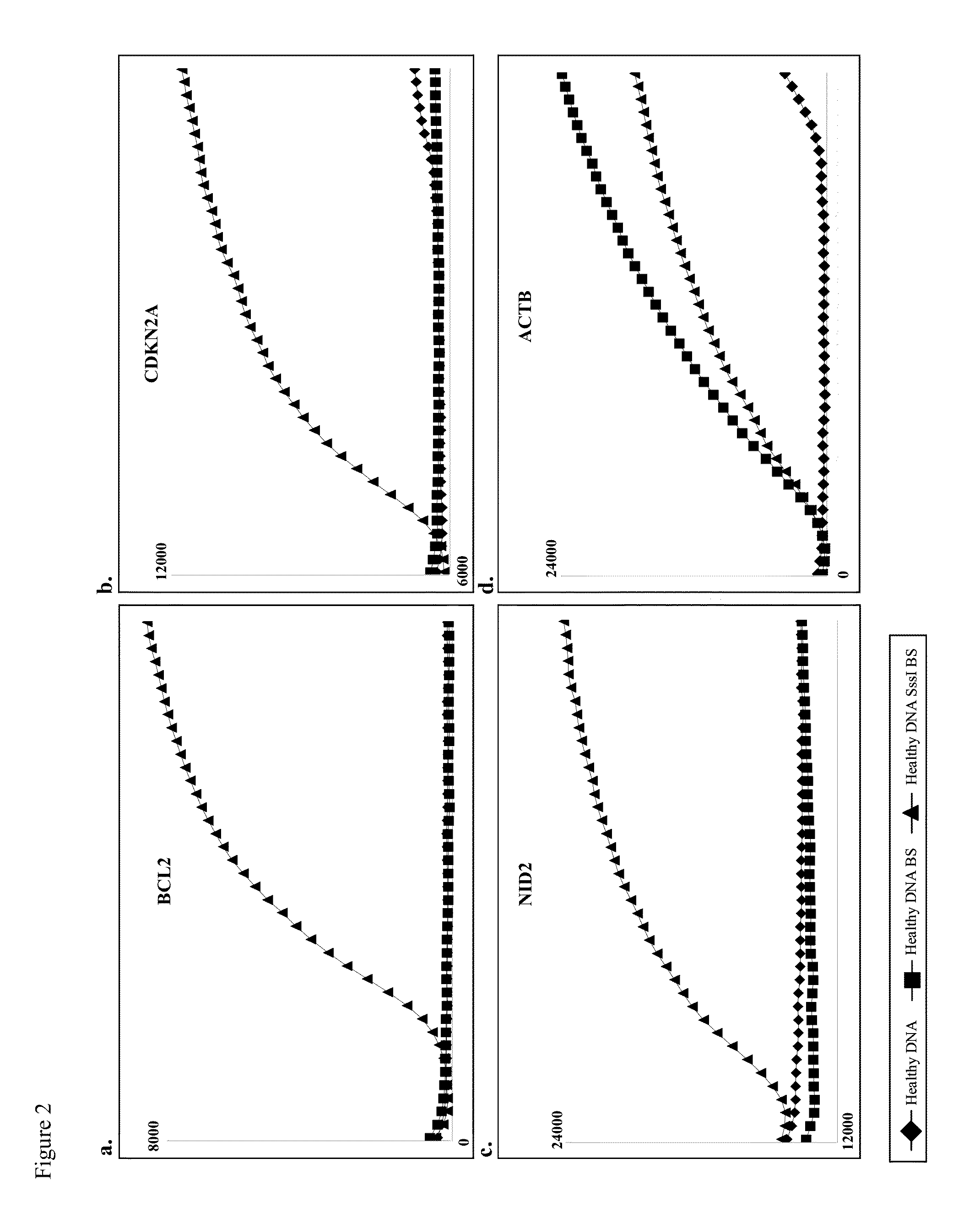
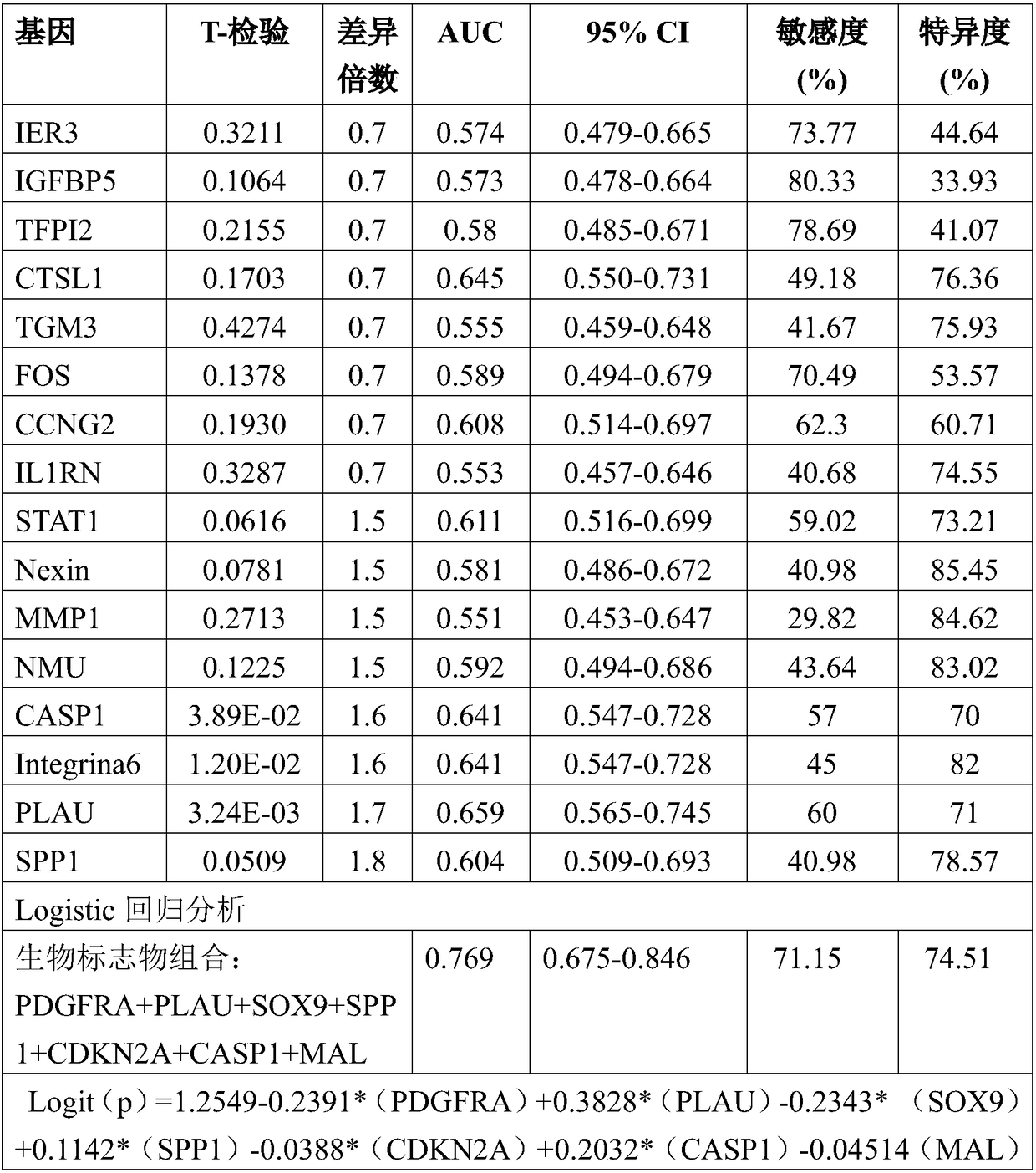
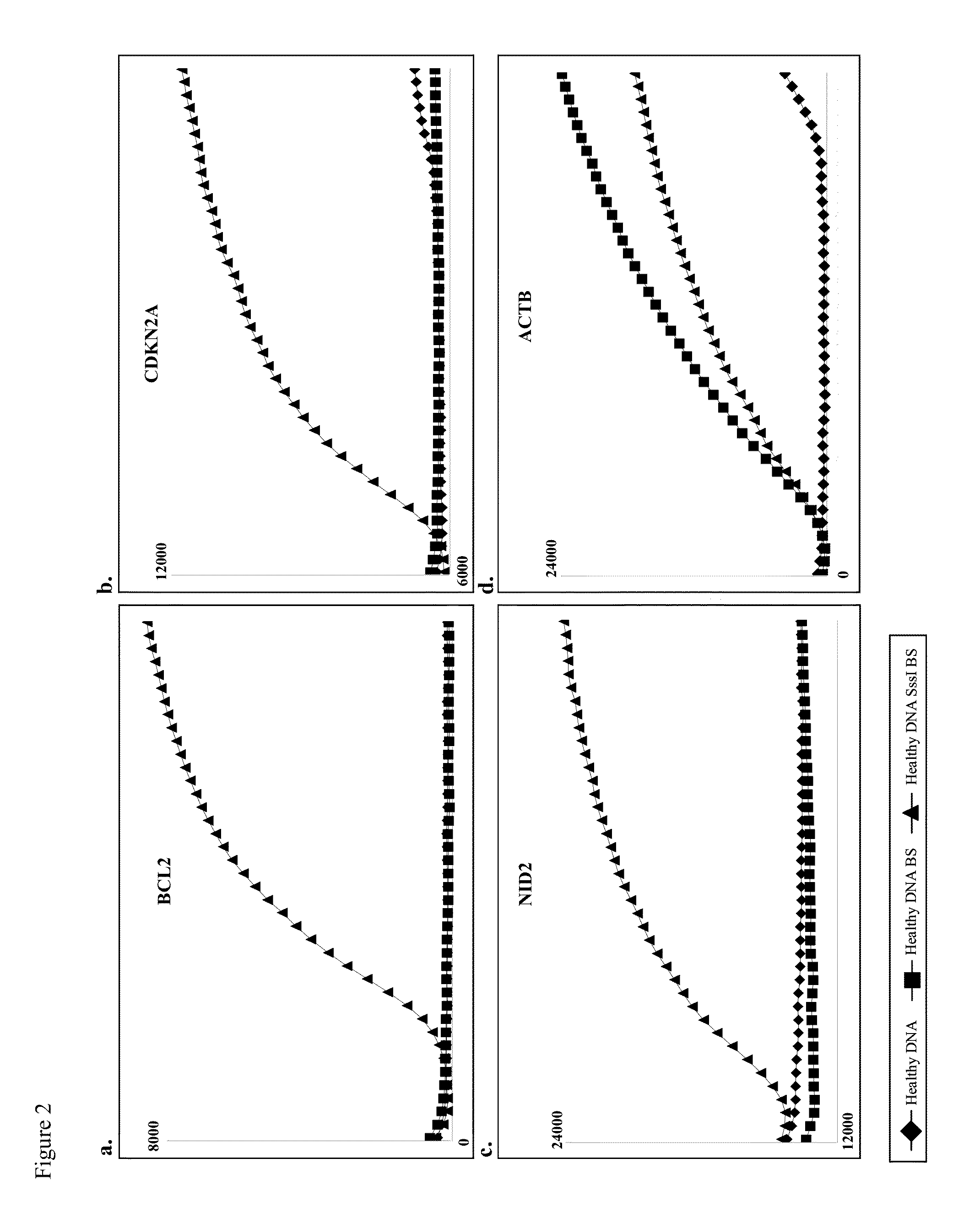
Detecting DNA methylation of BCL2, CDKN2a and NID2 genes to predict bladder cancer in humans
InactiveUS20130224738A1High riskEasy to detectMicrobiological testing/measurementDNA methylationBladder cancer
The present invention provides a method of detecting DNA methylation of a plurality of genes consisting of CDKN2A, BCL2 and NID2 in a urine sample from a human. Methods and compositions are provided herein for detecting and diagnosing bladder cancer by obtaining a urine sample from a human subject suspected of bladder cancer, followed by detecting DNA methylation of CDKN2A, BCL2 AND NID2 in urine samples from the individual. The present method permits specific detection of DNA methylation of the selected gene promoters in urine as a biomarker for bladder cancer in humans.
Owner:MEDICAL DIAGNOSTIC LAB
Kit for detecting susceptibility gene SNP locus of nasopharynx cancer
InactiveCN103045743AAchieve forecastRealize early warningMicrobiological testing/measurementNasopharyngeal carcinomaHigh risk group
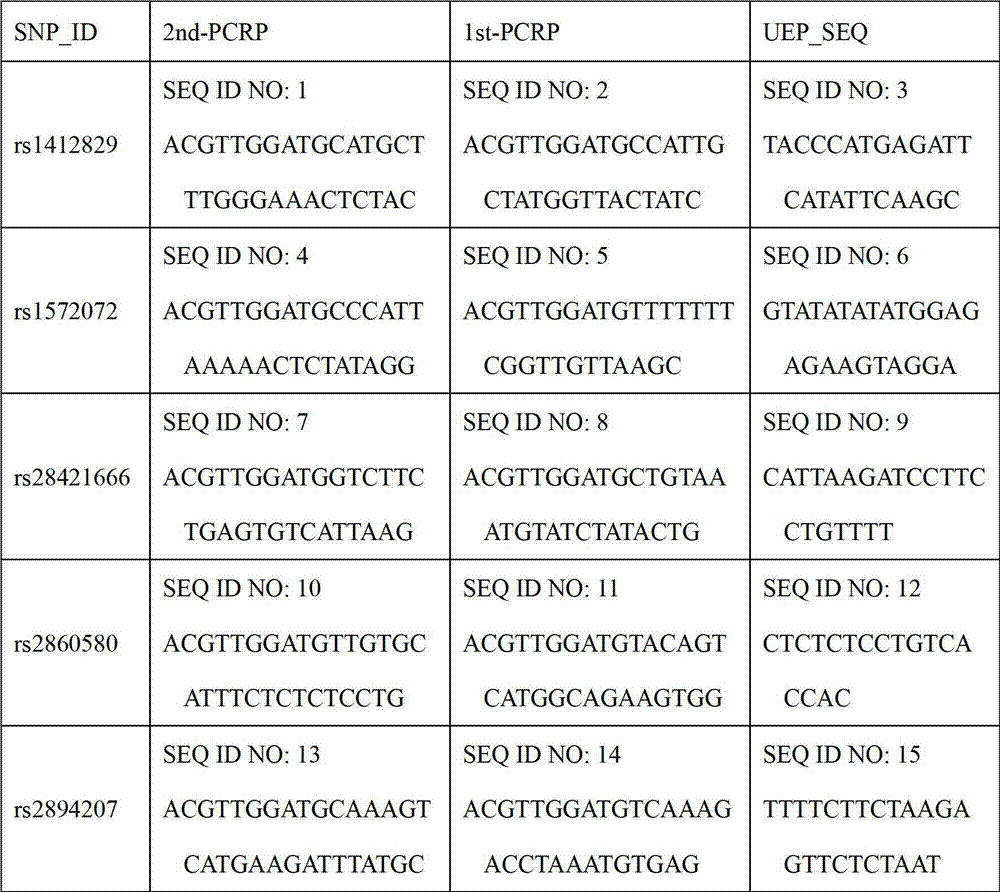
The invention relates to a kit for detecting susceptibility gene SNP locus of nasopharynx cancer, which is realized through the steps as follows: detecting a PCR amplimer and a single-base extension primer of an SNP locus rs2860580 of a susceptibility gene CDKN2A-CDKN2B of nasopharynx cancer; detecting PCR amplimers and single-base extension primers of SNP loci rs1572072 and rs9510787 of a susceptibility gene TNFRSF19 area of nasopharynx cancer; detecting PCR amplimers and single-base extension primers of SNP loci rs28421666,rs2860580 and rs2894207 of a susceptibility gene HLA of nasopharynx cancer; and detecting a PCR amplimer and a single-base extension primer of an SNP locus rs6774494 of a susceptibility gene MDS1-EVI1 of nasopharynx cancer. The kit provided by the invention can be used for simultaneously detecting the SNP loci of a plurality of susceptibility genes of nasopharynx cancer, and provides reference for estimating risk degree of individuals affected by nasopharynx cancer, general survey of populations with high nasopharynx cancer incidence, screening of high risk group attacked by nasopharynx cancer and implementation of relative precautionary measures.
Owner:SUN YAT SEN UNIV CANCER CENT
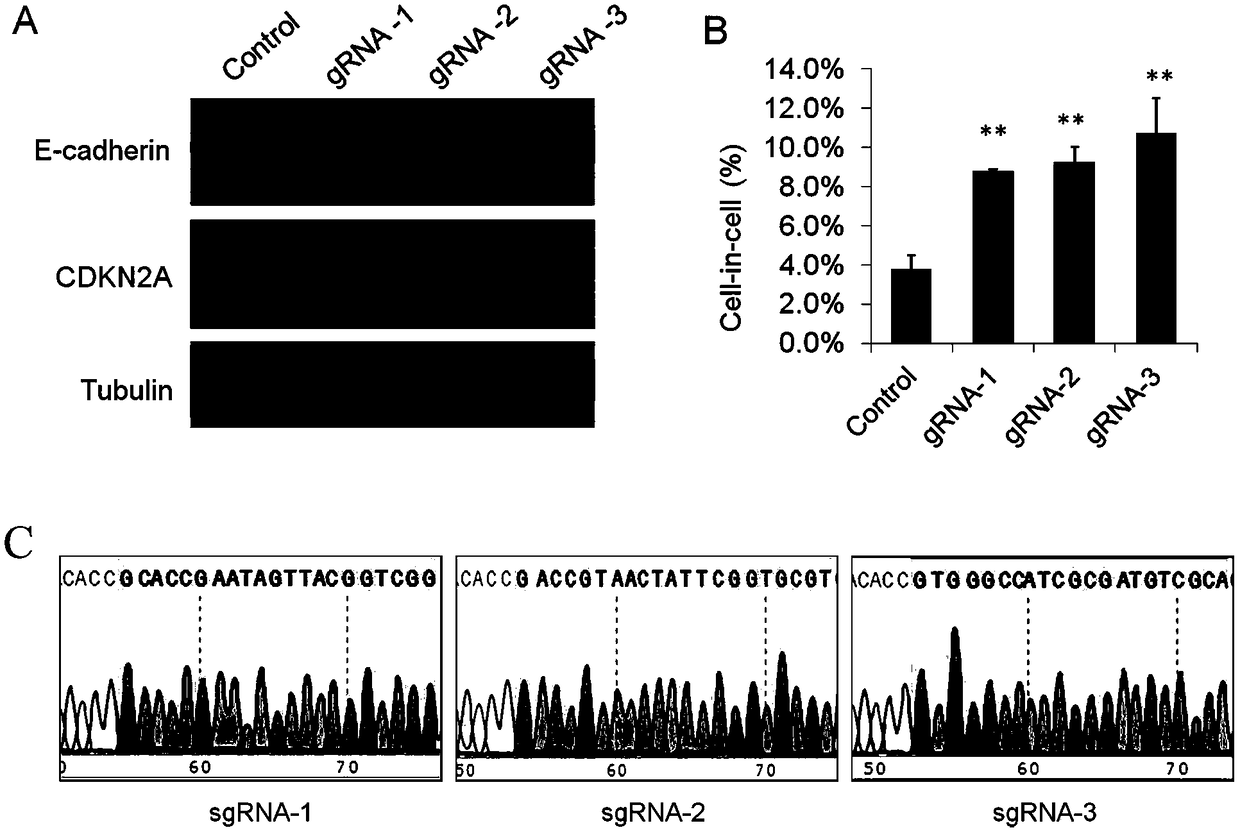
CIC cell models targeting to CDKN2A and preparation methods of CIC cell models
ActiveCN108588028APromote formationAccelerated deathMicrobiological testing/measurementPeptidesExpression geneTumor cells
The invention discloses cell-in-cell (CIC) cell models targeting to CDKN2A and preparation methods of the cell-in-cell (CIC) cell models. For the methods for preparing the cell-in-cell relevant cell models provided by the invention, the cell-in-cell relevant cell models are prepared by regulating the expression of the CDKN2A gene in the target cell. An inducing cell-in-cell cell model can be prepared by inhibiting the expression of the CDKN2A gene in the target cell, and an inhibiting cell-in-cell cell model can be prepared by realizing overexpression of the CDKN2A gene in the target cell. Forthe method for promoting the formation of the CIC structure by interfering the expression of the CDKN2A gene provided by the invention, the complete tumor cell is expressed aiming at CDKN2A, throughthe method, the intracellular death mediated by CIC can be promoted, and the purposes of killing tumor cells and inhibiting tumor growth can be realized.
Owner:ACADEMY OF MILITARY MEDICAL SCI
Diabetes type II gene susceptivity and pre-warning detection kit
The present invention provides a diabetes type II gene susceptivity and pre-warning detection kit, which is characterized in that seven nucleic acid (DNA and RNA) molecule markers are combined, wherein the seven nucleic acid (DNA and RNA) molecule markers comprise APM1 (+45), CDKAL1 (rs10946398), TCF7L2 (rs7903146) , CDKN2A / 2B (rs10811661), KCNQ1 (rs2237892), VDR (FokI) and SLC30A8 (rs13266634). The kit comprises: specific amplification primer pairs and DNA sequencing primer pairs of the gene polymorphism loci, PCR reaction liquids, PCR product enzymolysis liquids and DNA sequencing reaction liquids. According to the present invention, detection is performed by using the kit, gene information is processed and analyzed, diabetes type II susceptivity of a healthy subject is evaluated, and guiding significance is provided for personalized treatment and health care of diabetes type II people.
Owner:SHANGHAI JINFANG BIOLOGICAL TECH
Cardia adenocarcinoma auxiliary diagnosis kit related to a group of genes
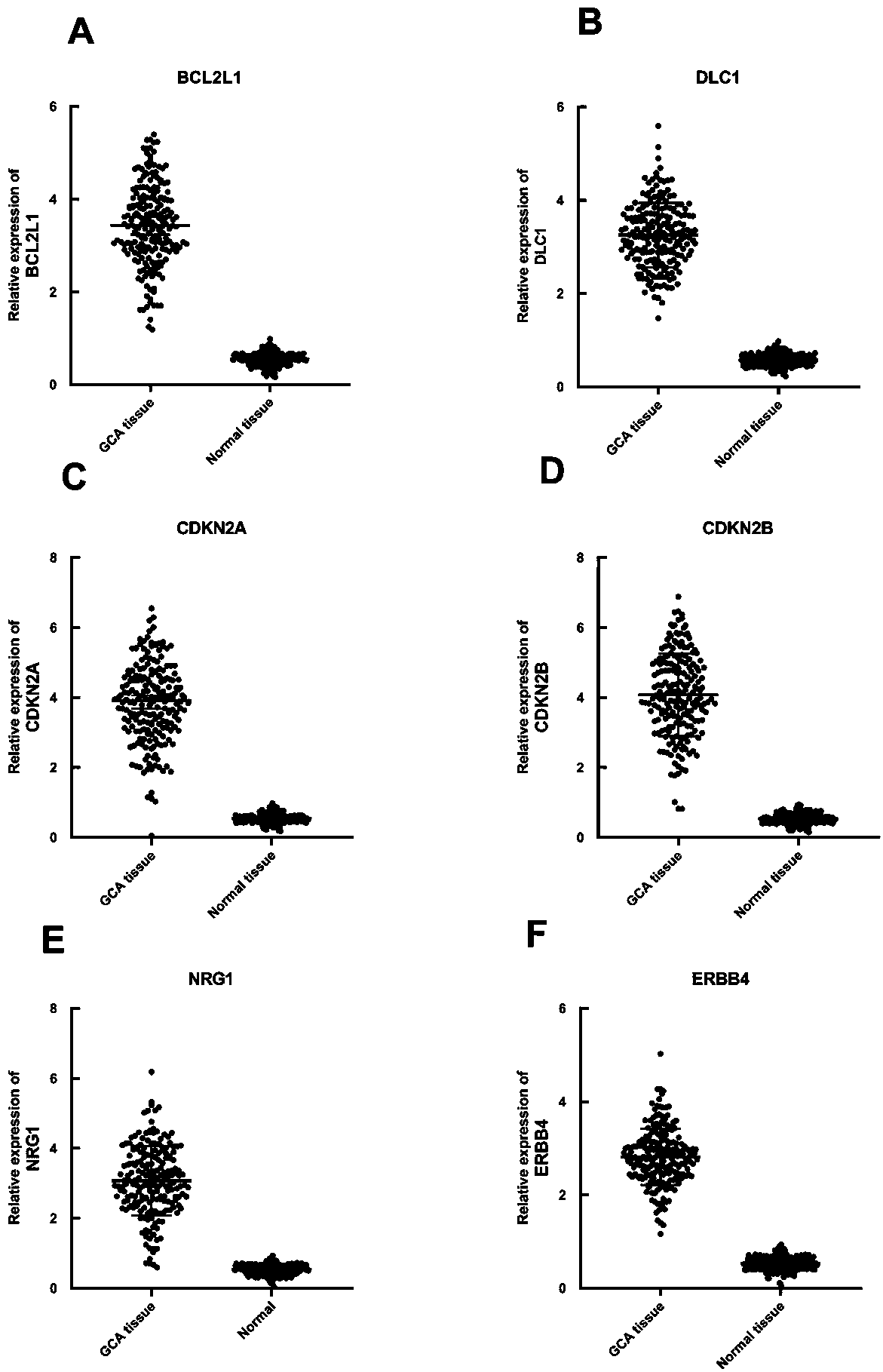
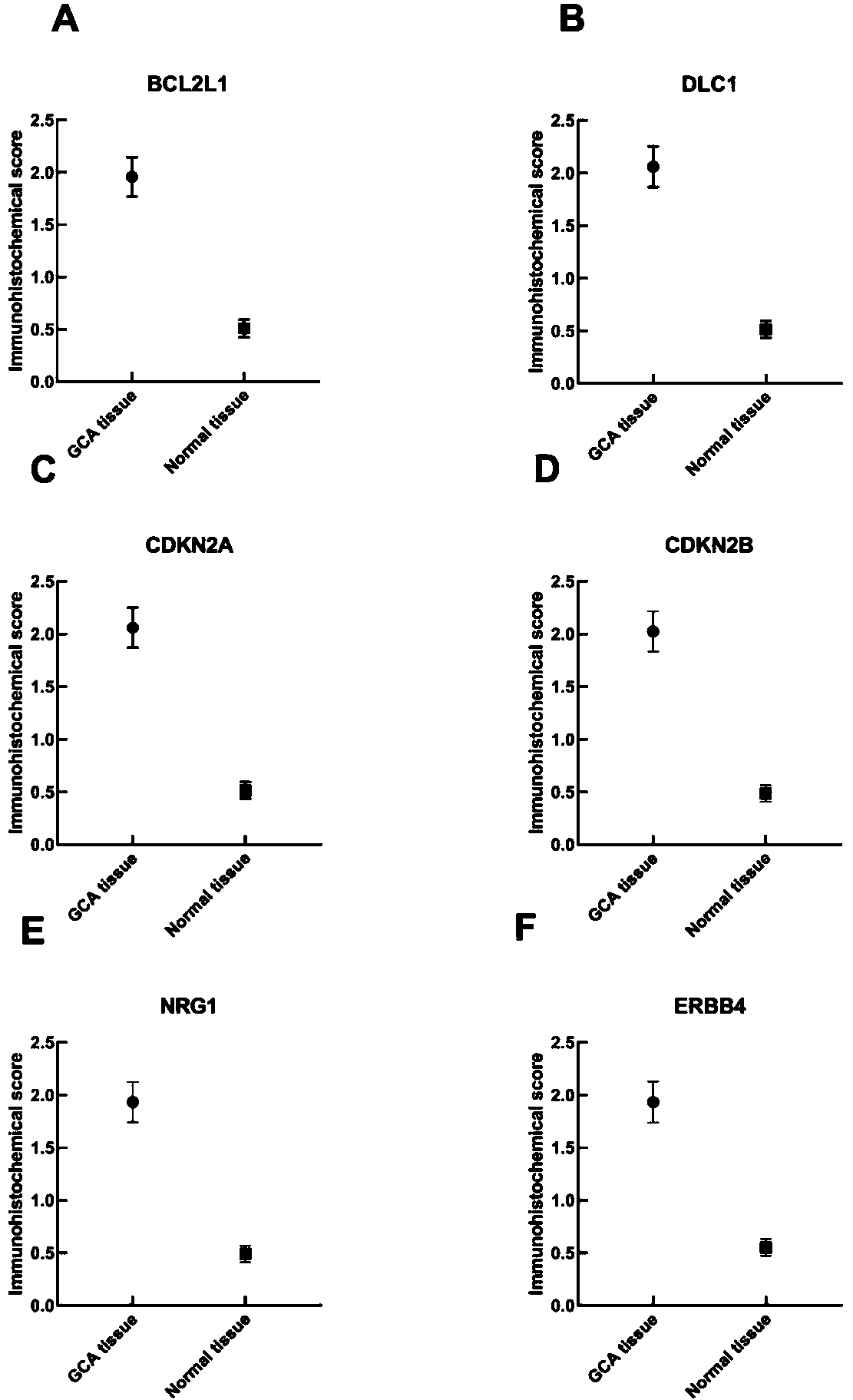
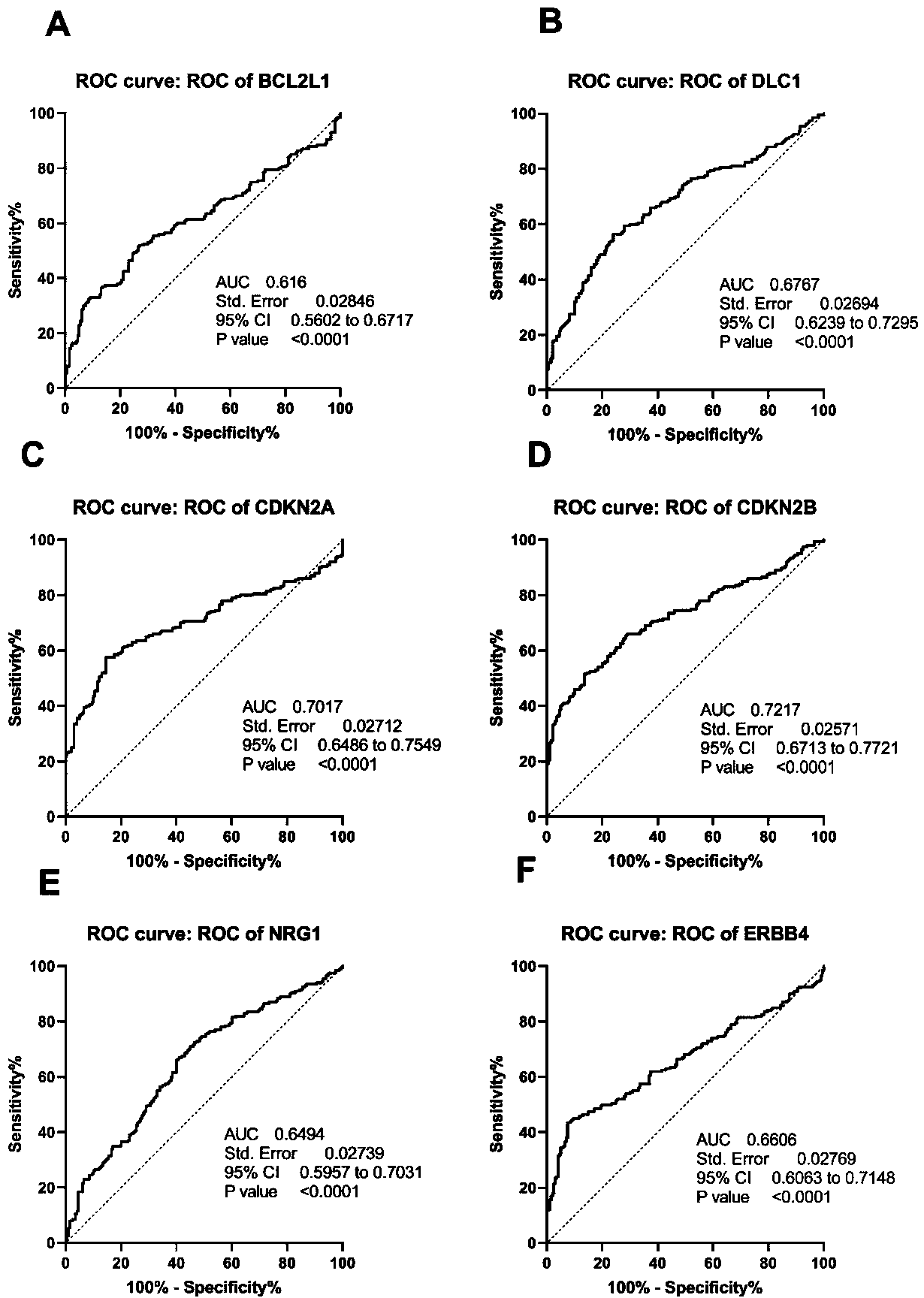
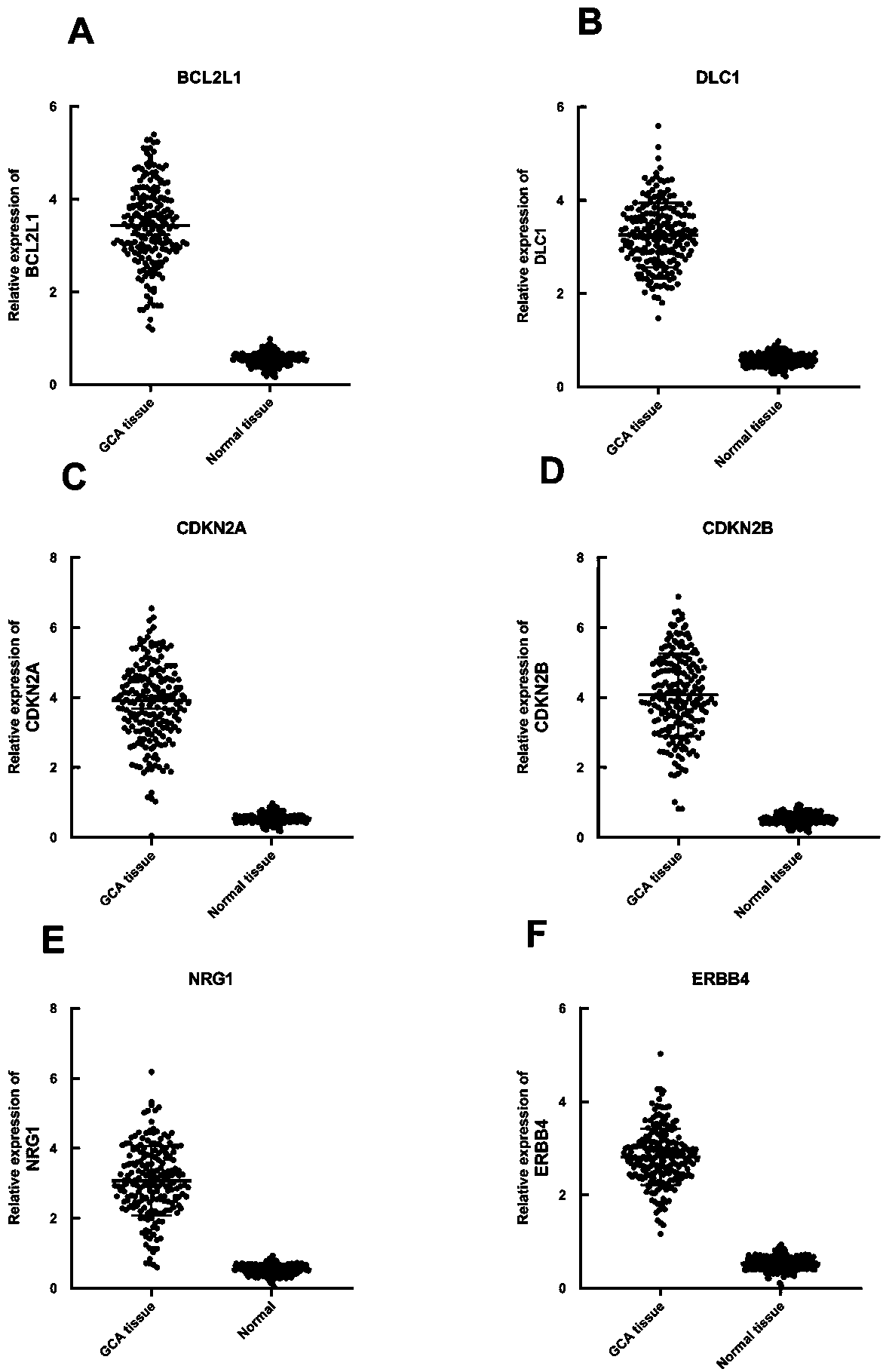
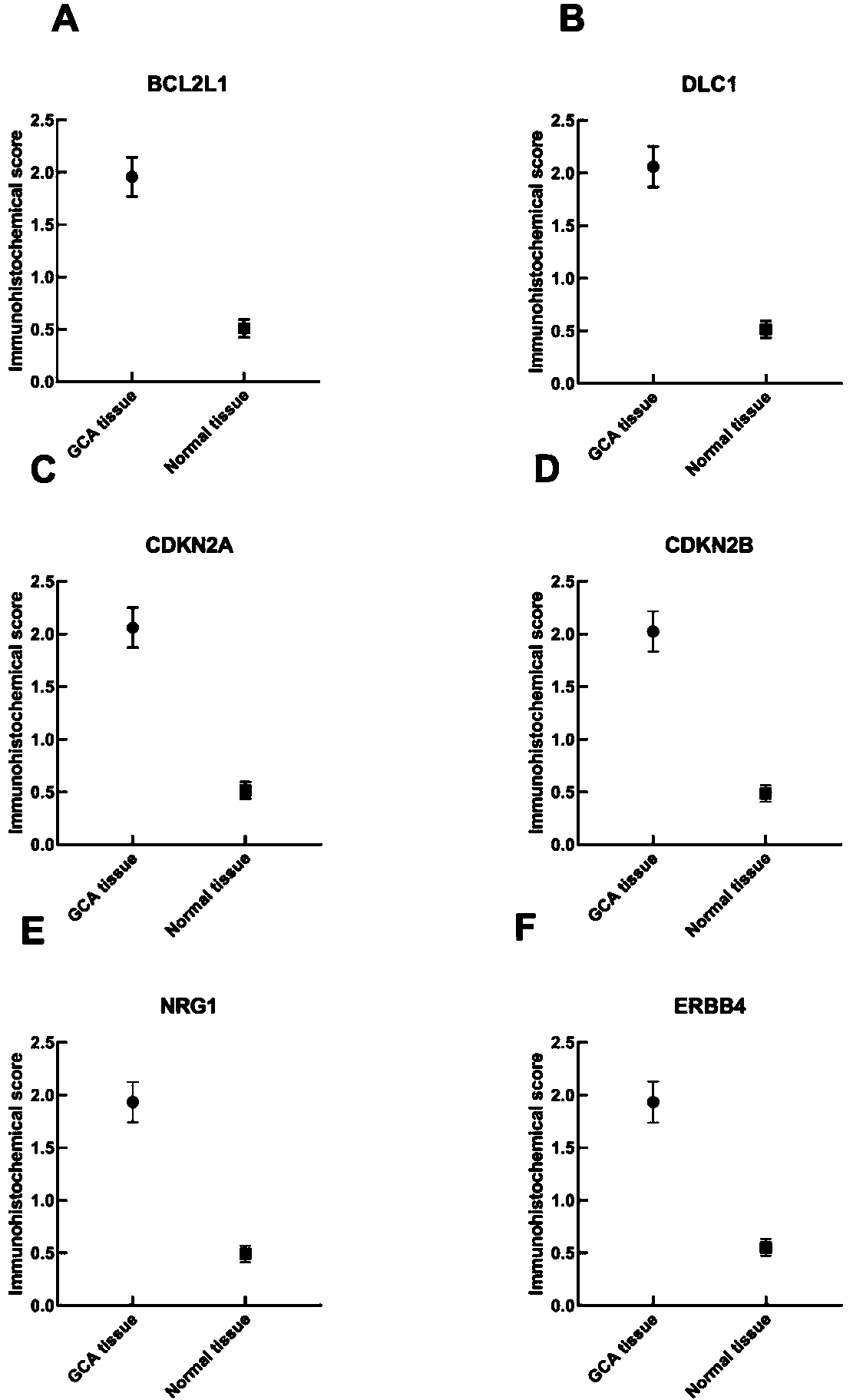
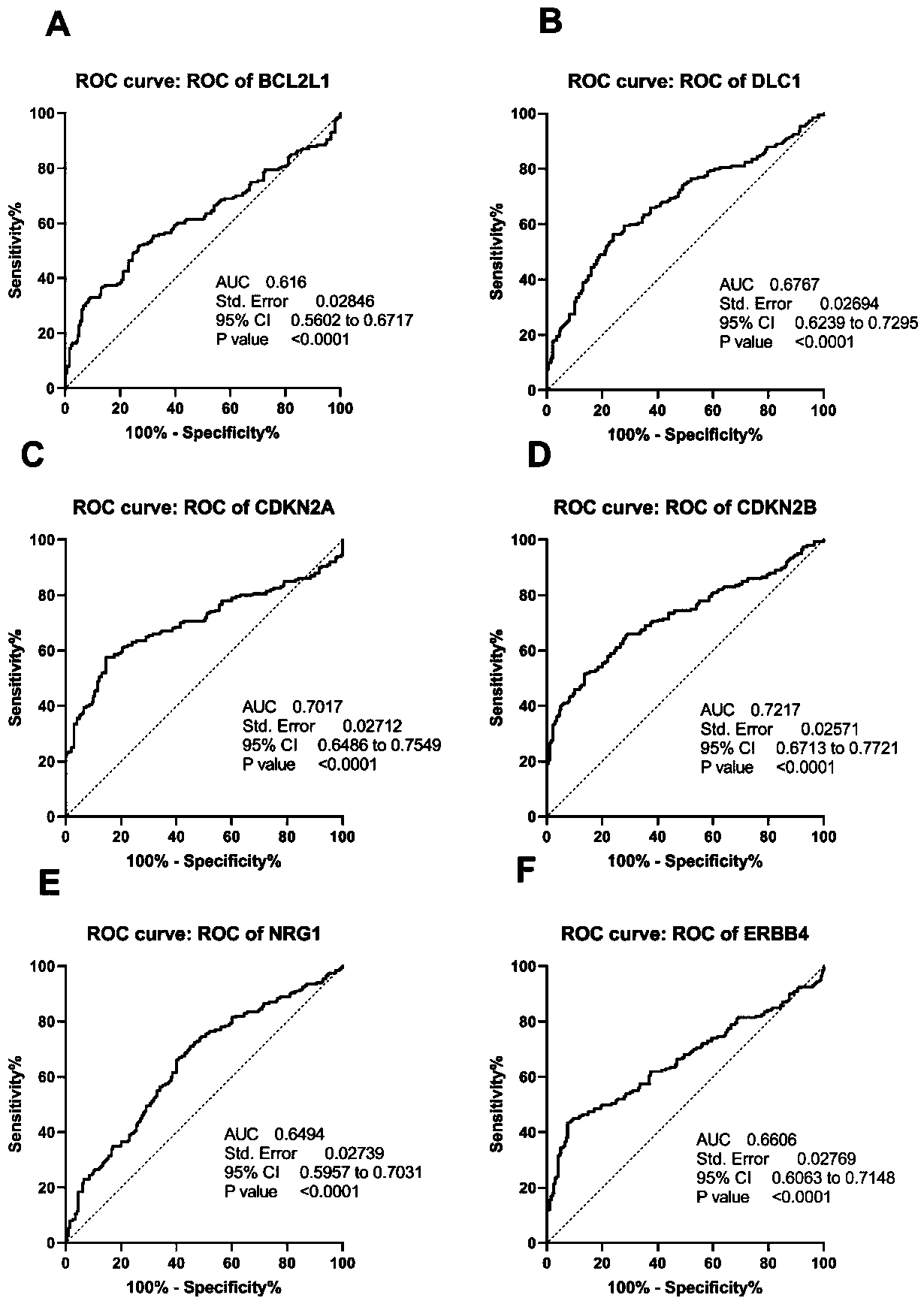
The invention relates to a cardia adenocarcinoma auxiliary diagnosis kit related to a group of genes, belongs to the technical field of biomedicine and molecular biology, and particularly relates to an ELISA kit for auxiliary diagnosis of cardia adenocarcinoma. The kit comprises a solid phase carrier and six antibodies which are respectively coated on the solid phase carrier, wherein the six antibodies are as follows: BCL2L1 antibody, DLC1 antibody, CDKN2A antibody, CDKN2B antibody, NRG1 antibody, ErbB4 antibody. Experiments prove that the group of related gene expression product has high expression in the serum of a cardia adenocarcinoma patient, the expression level of the related gene in the human serum is detected by the ELISA kit, and the detection result can be used for auxiliary diagnosis of the cardia adenocarcinoma such as early-onset screening, prognosis evaluation and the like, and the ELISA kit for auxiliary diagnosis of cardia adenocarcinoma has profound clinical significance and important popularization and application prospect.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
Kit for predicting recurrence and transference risk of patient suffering from early-stage operable esophageal squamous carcinoma
InactiveCN106498085AAccurate transferReduce transferMicrobiological testing/measurementWNT1Cyclin D1
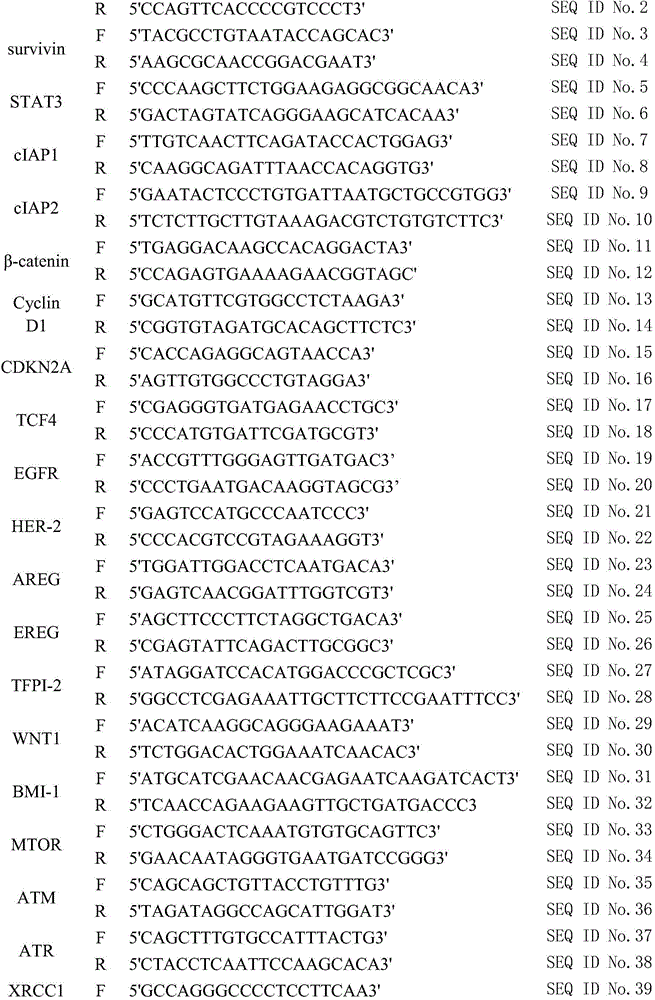
The invention provides a kit for predicting recurrence and transference risk of a patient suffering from early-stage operable esophageal squamous carcinoma. The kit contains amplification primers of 32 genes of BCL2, Survivin, STAT3, cIAP1, cIAP2, beta-catenin, Cyclin D1, CDKN2A, TCF4, EGFR, HER-2, AREG, EREG, TFPI-2, WNT1, BMI-1, MTOR, ATM, ATR, XRCC1, XRCC2, XRCC3, XRCC4, XRCC5, HIF-1alpha, HIF-2alpha, VEGF, RAD51, beta-actin, GAPDH, GUS and TFRC in all. The kit is capable of accurately predicting transference potential of early-stage operable esophageal squamous carcinoma, so that the transference and recurrence risk of the patient suffering from the early-stage operable esophageal squamous carcinoma after radical operation can be accurately predicted and evaluated, patients with high risk can be particularly monitored and effectively interfered, recurrence and transference of esophageal squamous carcinoma can be reduced, and prognosis of the patients can be further improved.
Owner:JIANGSU PROVINCE HOSPITAL
Combination of nelfinavir, metformin and rosuvastatin for treating cancer caused by aberrations in pten/tp53
The present disclosure relates to a method of treating cancer using a composition comprising Nelfinavir, Metformin, Rosuvastatin, optionally along with a pharmaceutically acceptable excipient. The said composition is used for the treatment of cancer caused due to aberration in PTEN gene, optionally along with aberration in TP53 gene and related genes selected from group comprising PI3K, CDKN2A, MDM2 and MDM4.
Owner:PRECISION PHARM INC
Gene markers for forecasting immunotherapy effect on cancer and application of gene markers
PendingCN112143810AImprove predictive performanceQuick filterMicrobiological testing/measurementTCF7L2Therapeutic effect
The invention discloses gene markers for forecasting an immunotherapy effect on a cancer and application of the gene markers. The markers comprise BRD4, RPS6KA4, TET1, BCL10, FGFR2, RAD51C, TCF7L2, TGFBR1, AR, BBC3, H3C11, TSC2, NSD1, PNRC1, IGF2, H3C3, SOCS1, TCF3, TSHR, PMS2, EGFR and CDKN2A. According to the application, a model constructed by 22 genes can be used for predicting an immunotherapy effect and better instructing clinical medication, and the susceptibility and specificity of prediction of the model are superior to those of PD-L1 expression.
Owner:TCM INTEGRATED HOSPITAL OF SOUTHERN MEDICAL UNIV
Kit for detecting mutation of lung adenocarcinoma cell cycle progress pathway related genes
ActiveCN113151479AEffectively guide individualized treatmentAvoid wastingMicrobiological testing/measurementMolecular diagnostic techniquesPredictive methods
The invention relates to the technical field of molecular diagnosis, in particular to a composition for detecting mutation of lung adenocarcinoma cell cycle progress pathway related genes (CDKN2A, CCND1, CCNE1, CDK4 and RB1) and application of the composition, and the composition is composed of reagents for detecting the expression quantity of the related genes. RNA sequencing, LASSO and binary classification Logistic regression are applied to screen genes related to the cell cycle progress pathway, a score Score is constructed, a cut-off value corresponding to the Score in the lung adenocarcinoma is obtained through ROC analysis, and the method can be used for predicting mutation of the lung adenocarcinoma cell cycle progress pathway related genes. The specific gene mutation of the lung adenocarcinoma is predicted by using the expression quantity of the gene composition, and verification shows that the prediction method has the advantages of high accuracy and good specificity, and has a good application prospect.
Owner:ZHONGSHAN HOSPITAL FUDAN UNIV
CDKN2A epitope peptide for detecting cervical cancer markers and application of CDKN2A epitope peptide
ActiveCN104558147AHigh expressionGuide early diagnosisTumor rejection antigen precursorsTumor specific antigensSerum igeEpitope
The invention relates to a CDKN2A epitope peptide for detecting cervical cancer markers and an application of the CDKN2A epitope peptide, and belongs to the technical field of biology. An amino acid sequence of the CDKN2A epitope peptide for detecting the cervical cancer markers is as shown in SEQ ID NO:1; and the CDKN2A epitope peptide for detecting the cervical cancer markers is applied to preparation of an early diagnosis kit for cervical cancer. The CDKN2A epitope peptide has the advantages that the autoantibody expression level in serum and plasma of a patient with the cervical cancer is detected and a corresponding reagent is developed; the dangerousness of the cervical cancer is predicted; and a foundation is laid for preparation of the early diagnosis kit for the cervical cancer.
Owner:刘林林
Detection kit for oral squamous cell carcinoma lymph node metastasis prediction
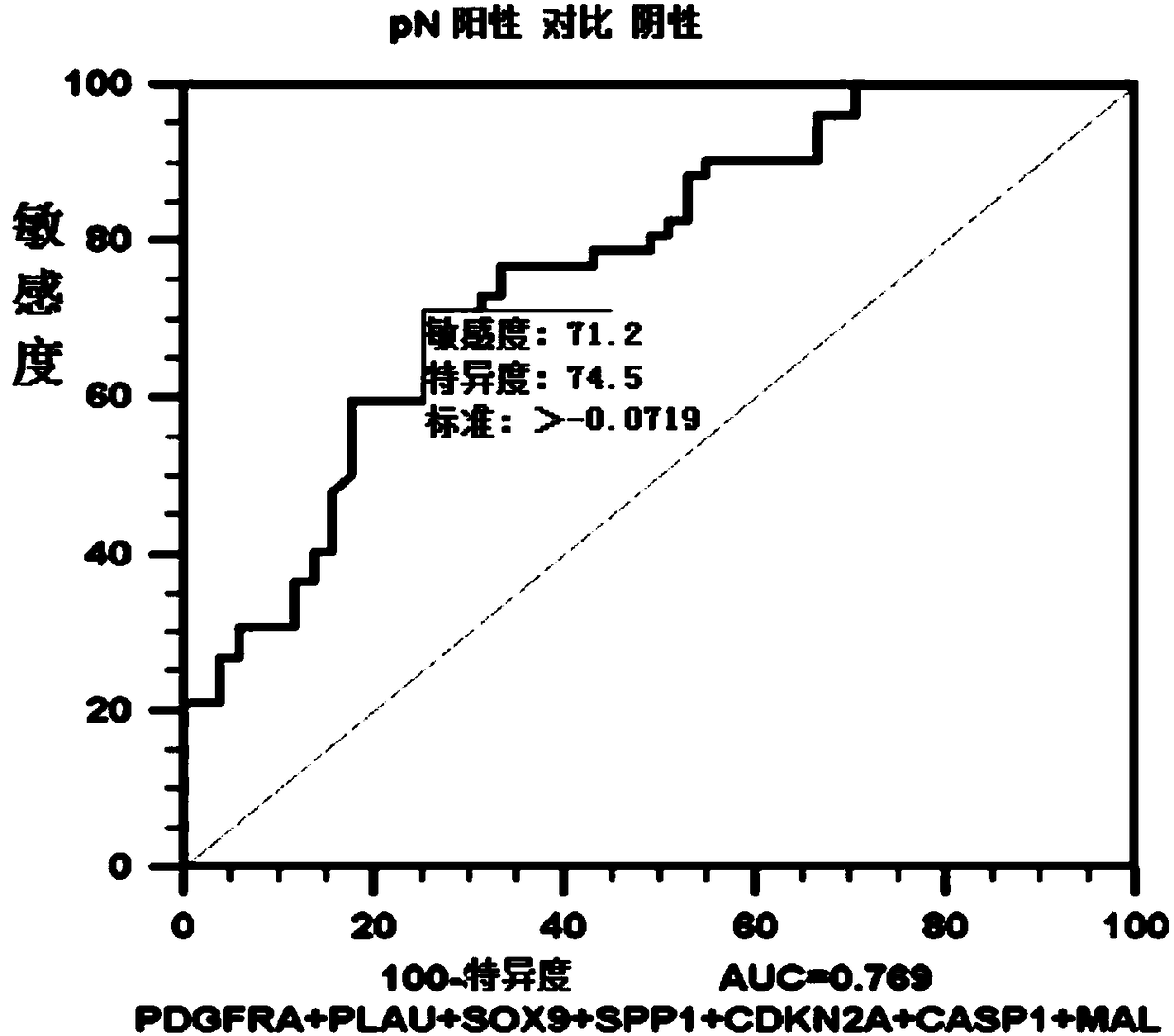
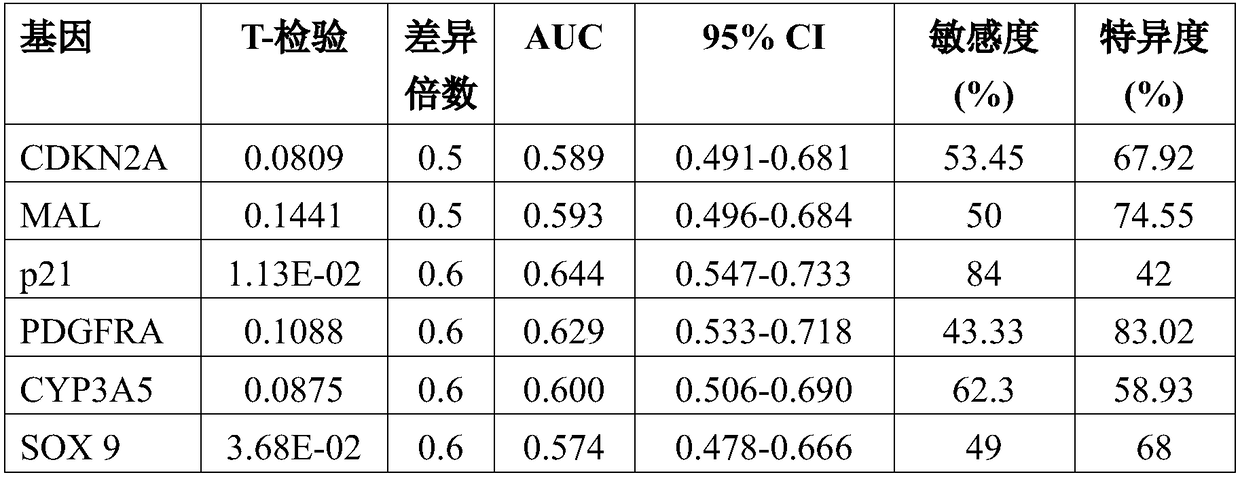
ActiveCN108546761AObjective predictionAccurate predictionMicrobiological testing/measurementForward primerSquamous Carcinomas
The invention relates to the technical field of gene detection, and discloses a detection kit for oral squamous cell carcinoma lymph node metastasis prediction. The kit comprises forward primer and reverse primer sequences of 7 kinds of genes including PDGFRA, PLAU, SOX9, SPP1, CDKN2A, CASP1 and MAL. The invention also discloses a method for screening the gene or the gene combination for the oralsquamous cell carcinoma lymph node metastasis prediction. The detection kit can be used for objectively, accurately and sensitively predicting the oral squamous cell carcinoma lymph node metastasis condition, so that the survival rate of the oral squamous cell carcinoma patients can be favorably improved.
Owner:SHANGHAI NINTH PEOPLES HOSPITAL AFFILIATED TO SHANGHAI JIAO TONG UNIV SCHOOL OF MEDICINE
Detecting DNA methylation of BCL2, CDKN2A and NID2 genes to predict bladder cancer in humans
InactiveUS9096905B2High riskEasy to detectMicrobiological testing/measurementBacteriuriaDNA methylation
The present invention provides a method of detecting DNA methylation of a plurality of genes consisting of CDKN2A, BCL2 and NID2 in a urine sample from a human. Methods and compositions are provided herein for detecting and diagnosing bladder cancer by obtaining a urine sample from a human subject suspected of bladder cancer, followed by detecting DNA methylation of CDKN2A, BCL2 AND NID2 in urine samples from the individual. The present method permits specific detection of DNA methylation of the selected gene promoters in urine as a biomarker for bladder cancer in humans.
Owner:MEDICAL DIAGNOSTIC LAB
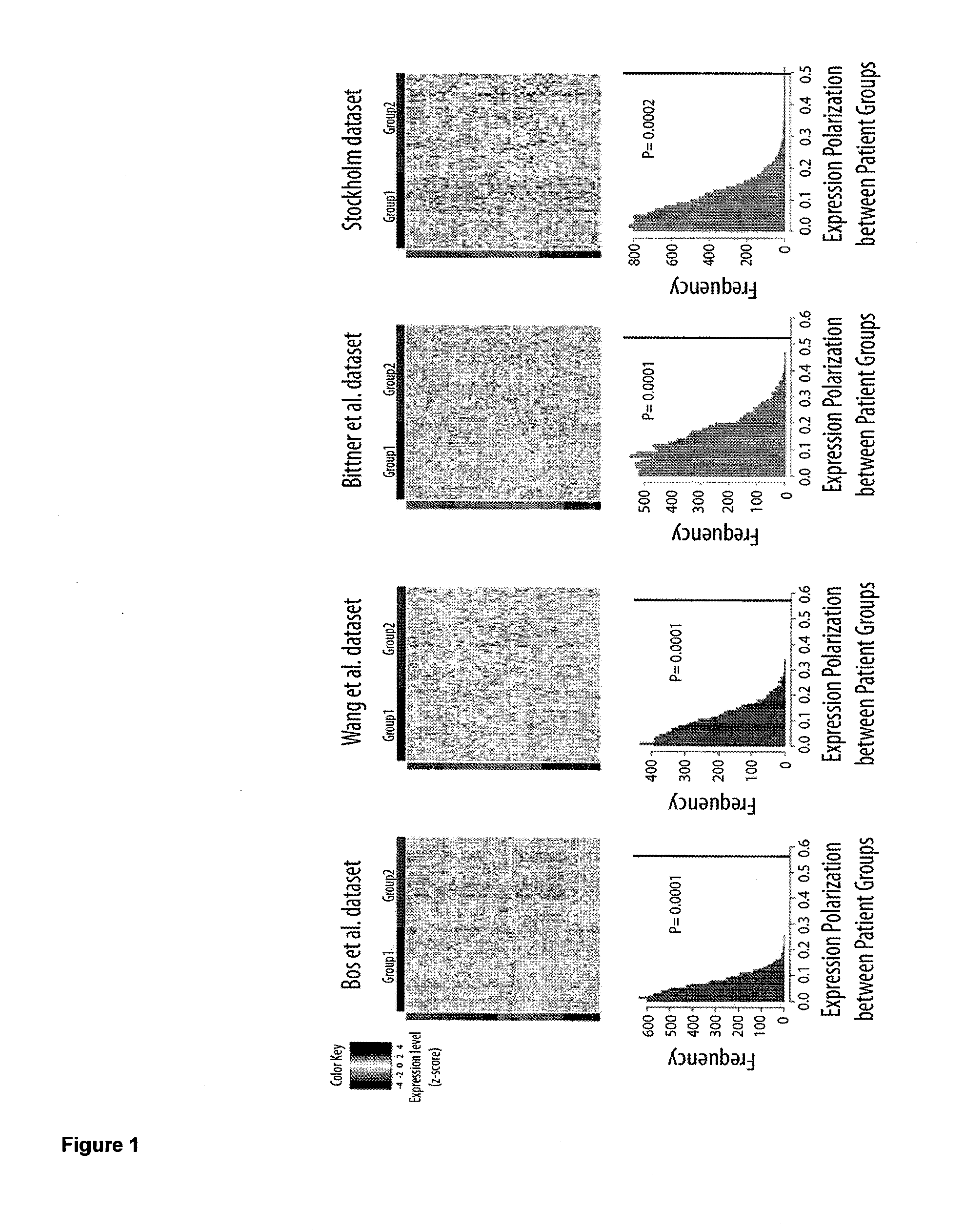
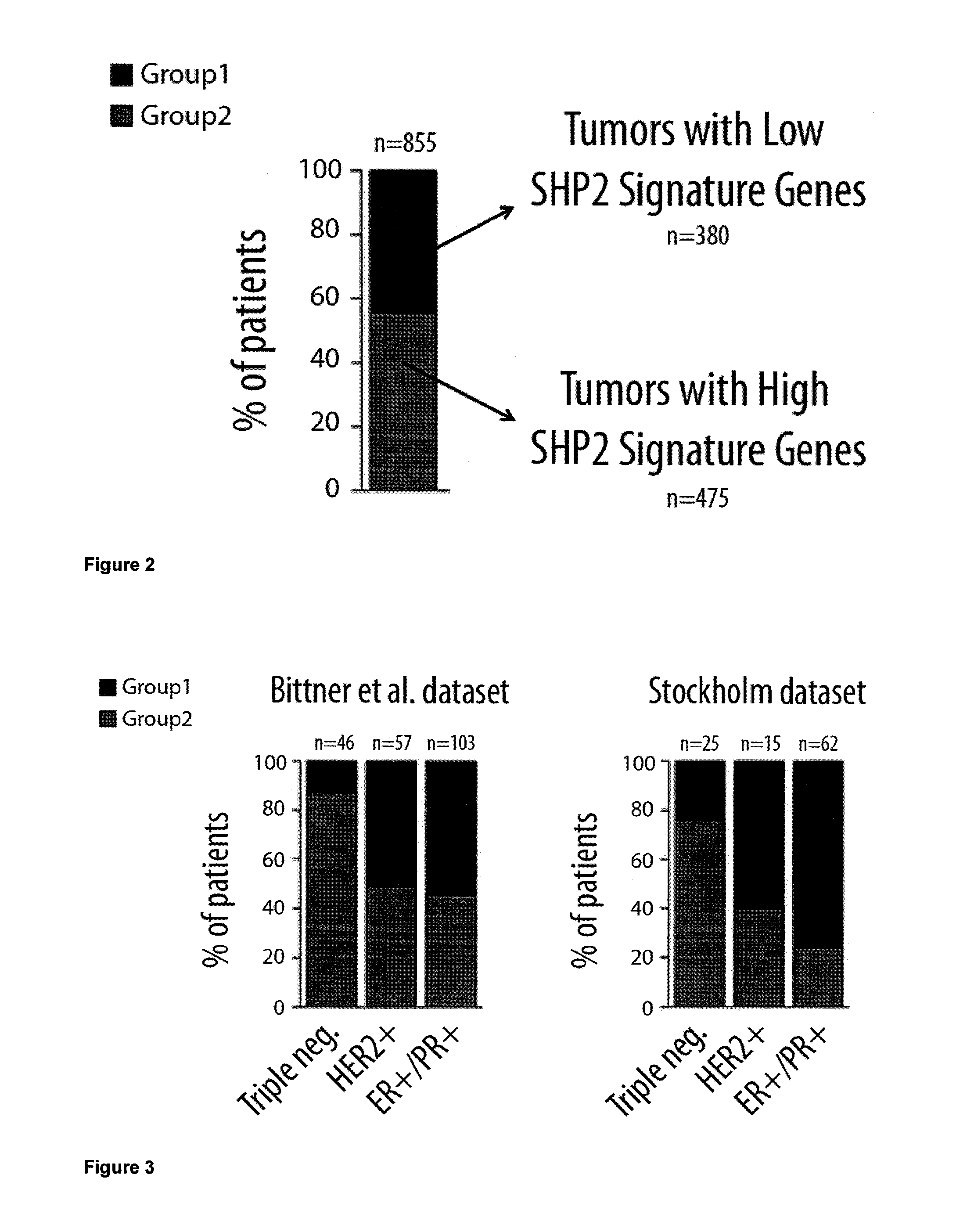
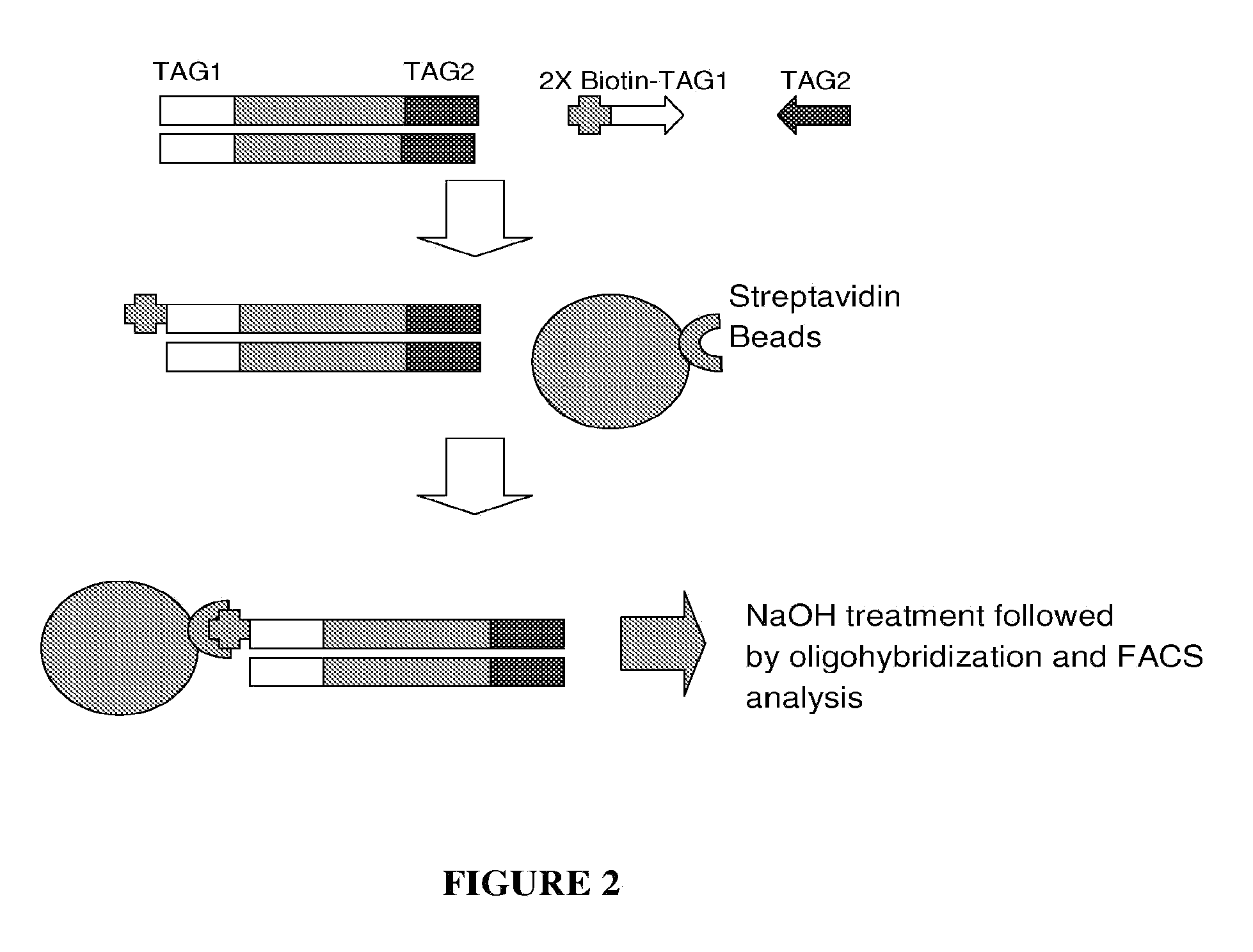
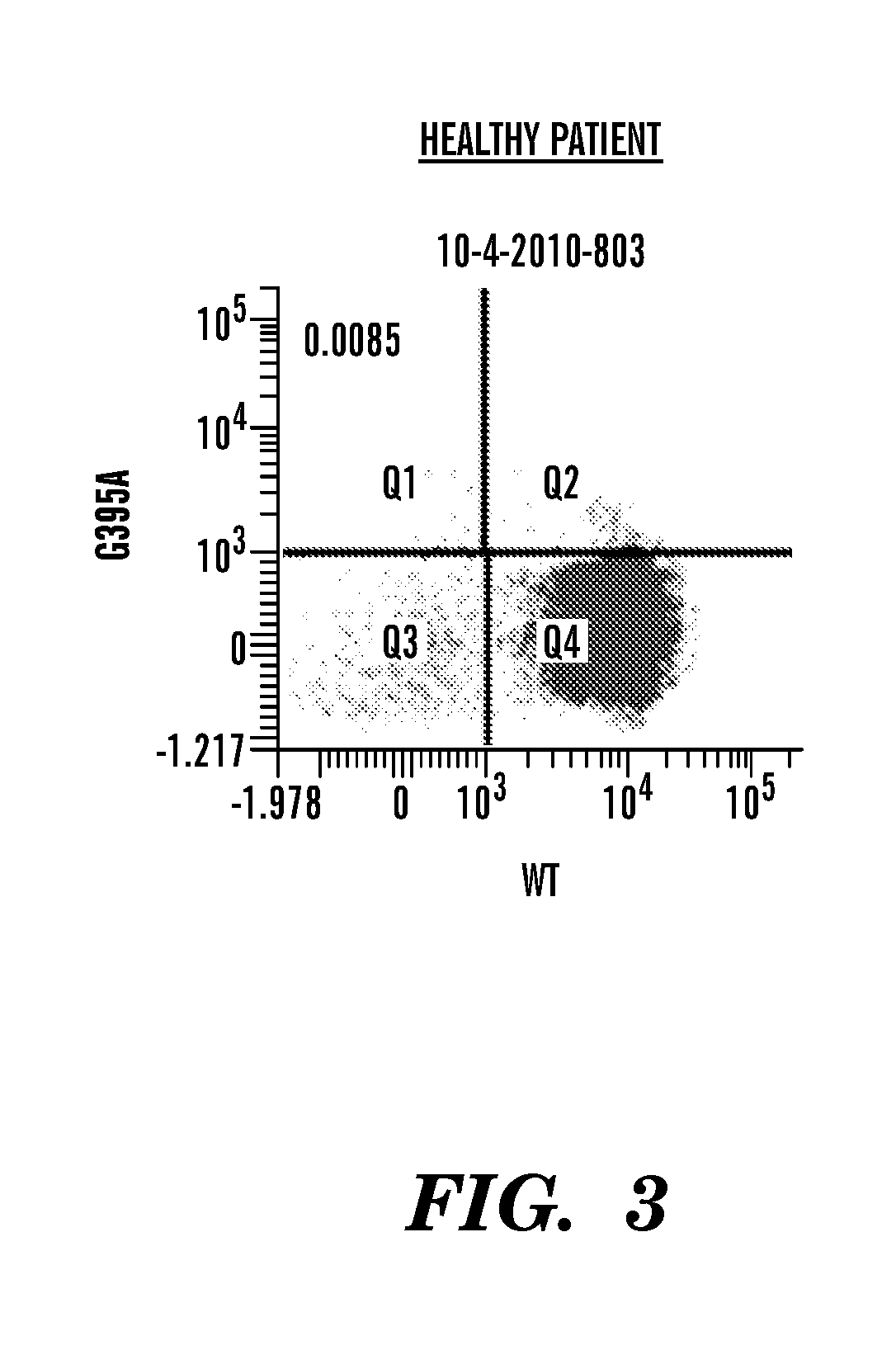
Protein tyrosine phosphatase, non-receptor type 11 (ptpn11) and triple-negative breast cancer
The present invention relates to a method for treating breast cancer in a subject having a breast cancer of the triple-negative type, which method comprises the step of administering to said subject a therapeutically effective amount of a modulator of the protein tyrosine phosphatase, non-receptor type 11 (PTPN11) gene or of its gene product (Shp2). The present invention also relates to a method for treating breast cancer in a subject having a breast cancer over-expressing the “SHP2 signature” genes, as compared to normal breast tissue samples, which method comprises the step of administering to said subject a therapeutically effective amount of a modulator of the protein tyrosine phosphatase, non-receptor type 11 (PTPN11) gene or of its gene product (Shp2), wherein said “SHP2 signature” genes consist of the genes SGCB, ZSCAN12, ID4, ZIC3, CPVL, HLA-A, MCOLN3, SPATA18, TMEM45A, GNAL, CYBRD1, TSPAN7, ZEB1, CNTLN, NEFL, CENPV, ARL6, HPRT1, LRRC34, PDPN, BEND7, SLC16A10, FAM27E1, PLEKHA1, HERC5, CHIC1, PHF6, ELOVL4, ANTXR1, PRAME, SCML1, CLIP4, CECR2, CNOT10, IGF2BP3, NAP1L3, GPC3, KIAA1804, DGKE, FAS, EPHA6, KDELC1, CRISPLD1, DOCK3, ACSL4, CNTNAP3, PLEKHM3, RDX, TBX18, RRAGD, HOXB5, SNCA, FUNDC2, ITGA8, HFM1, IGF2BP2, CCND2, SGTB, MKX, CRYBG3, WBP5, LPHN3, BEX4, CPNE8, GLDC, SLC35F1, HOXA13, SERPINF1, NEFM, SYCP2L, FHL1, APOBEC3C, CALD1, FKBP10, HOXD11, DENND2C, LRRC49, FAM55C, KIAA0408, HOXB9, C160RF62, ACN9, TUSC3, ELOVL2, SPOCK3, HOXB6, WDR35, MPP1, FBX038, PRKAA2, SLAIN1, NPHP3, KIAA1524, PRPS1, GJC1, AMOT, SLC9A6, KCTD12, NUP62CL, DZIP3, JAM3, HOXA9, ANKRD19, CDKN2A, BCAT1, OAT, LPHN2, CCDC82, HSD17B11, SAMHD1, WDR17, STK33, GSTP1, TRPC1, CKB, LIN28B, ALDH1L2, SACS, CLGN, MY03A, EPB41L3, SLC25A27, VCAN, GPX8, GALNT13, PVRL3, MOXD1, HEY1, MAP7D3, ESD, MPP6, EYA4, SPG20, ZDBF2, ZNF204, IFT57, AKR1B1, ADAT2, ZNF717, CCDC88A, ZNF215, MIDI, FBN2, LOC100130876, TCEAL8, IGF2BP1, ANKRD18B, PLAGL1, PM20D2, LDHB, C150RF51, PTPN11, EPB41L2, TLE4, GOLM1, C60RF192, HOXD13, SLIT2, UCHL1, DYNC2H1, CPS1, GPR180, PYGL, NRN1, PRTFDC1, SLC16A1, DSC3, TMC01, LRCH2, SLC6A15, DZIP1, HOXA5, HSPA4L, CDR1, PLS3, ECHDC1, SMARCA1, CXORF57, HOXD10, and IRS4.
Owner:NOVARTIS FORSCHUNGSSTIFTUNG ZWEIGNIEDERLASSUNG FRIEDRICH MIESCHER INSTITTUE FOR BIOMEDICAL RES
Microvesicle-based assays
Methods are disclosed herein for assaying a biological sample or a bodily fluid obtained from a subject by isolating, obtaining or using a microvesicle fraction from the biological sample or bodily fluid and detecting in the microvesicle fraction the presence or absence of a genetic aberration in an IDH1, TDH2, TP53, PTEN, CDKN2A, NF1, EGFR, RB1, PIK3CA, or BRAF gene. The methods may be used for aiding the diagnosis, prognosis, monitoring, or therapy selection in relation to a disease or other medical condition (e.g., a glioma) in a subject.
Owner:THE GENERAL HOSPITAL CORP
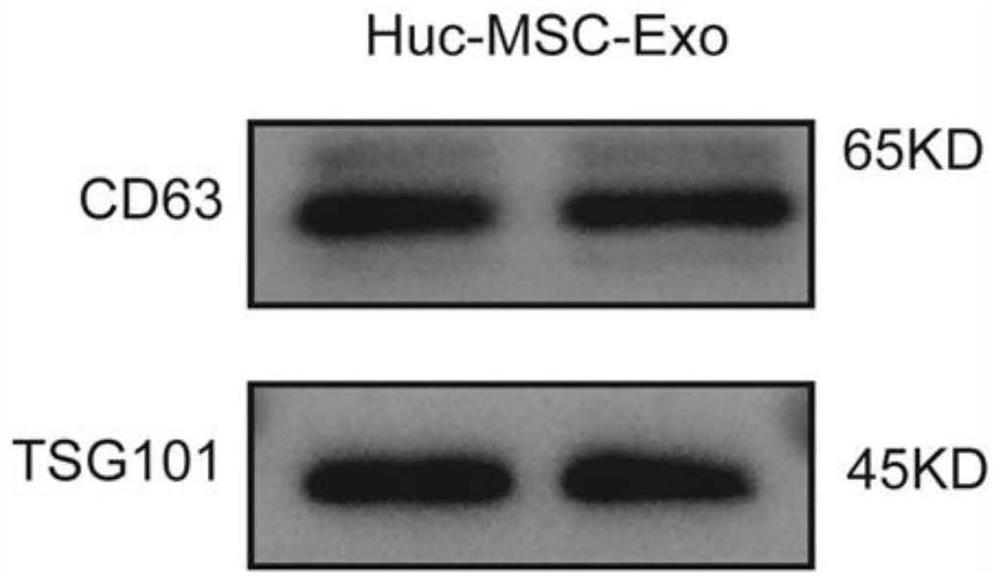
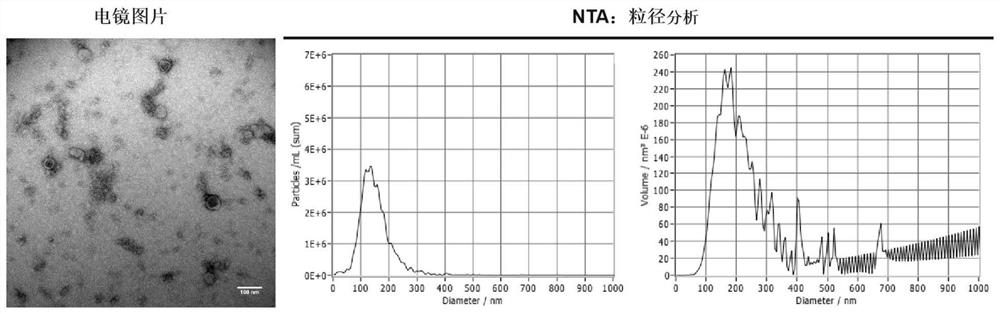
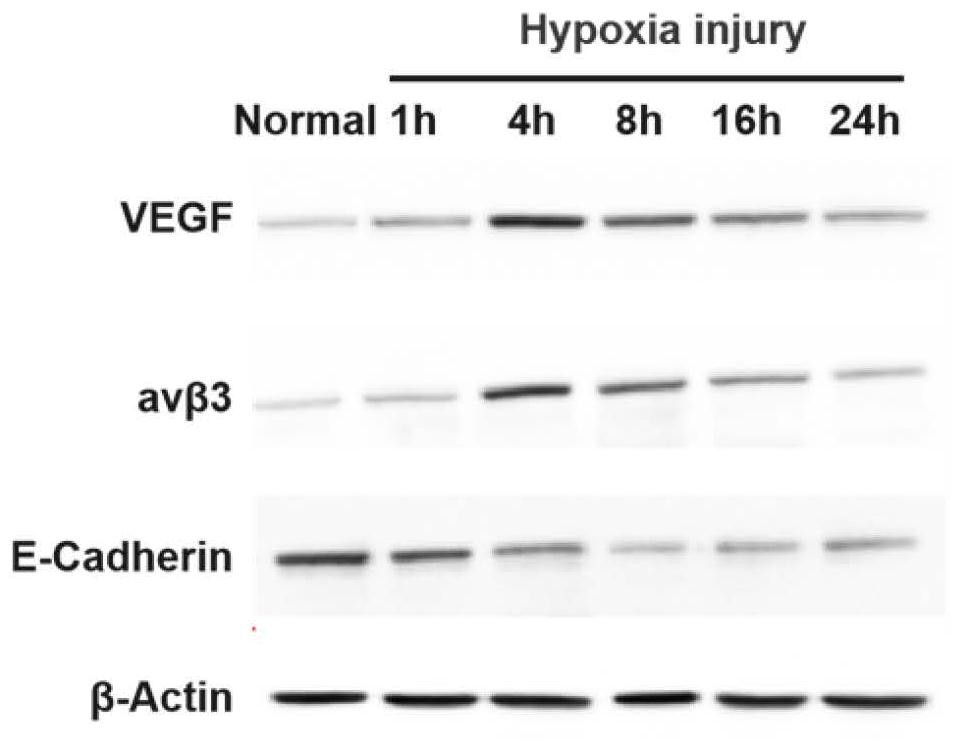
Application of miR-663a or target gene CKDN2A thereof in thin endometrium
PendingCN114657241AInhibition of hypoxic injuryMicrobiological testing/measurementDisease diagnosisWestern blotEndostelium
Owner:PEKING UNION MEDICAL COLLEGE HOSPITAL CHINESE ACAD OF MEDICAL SCI
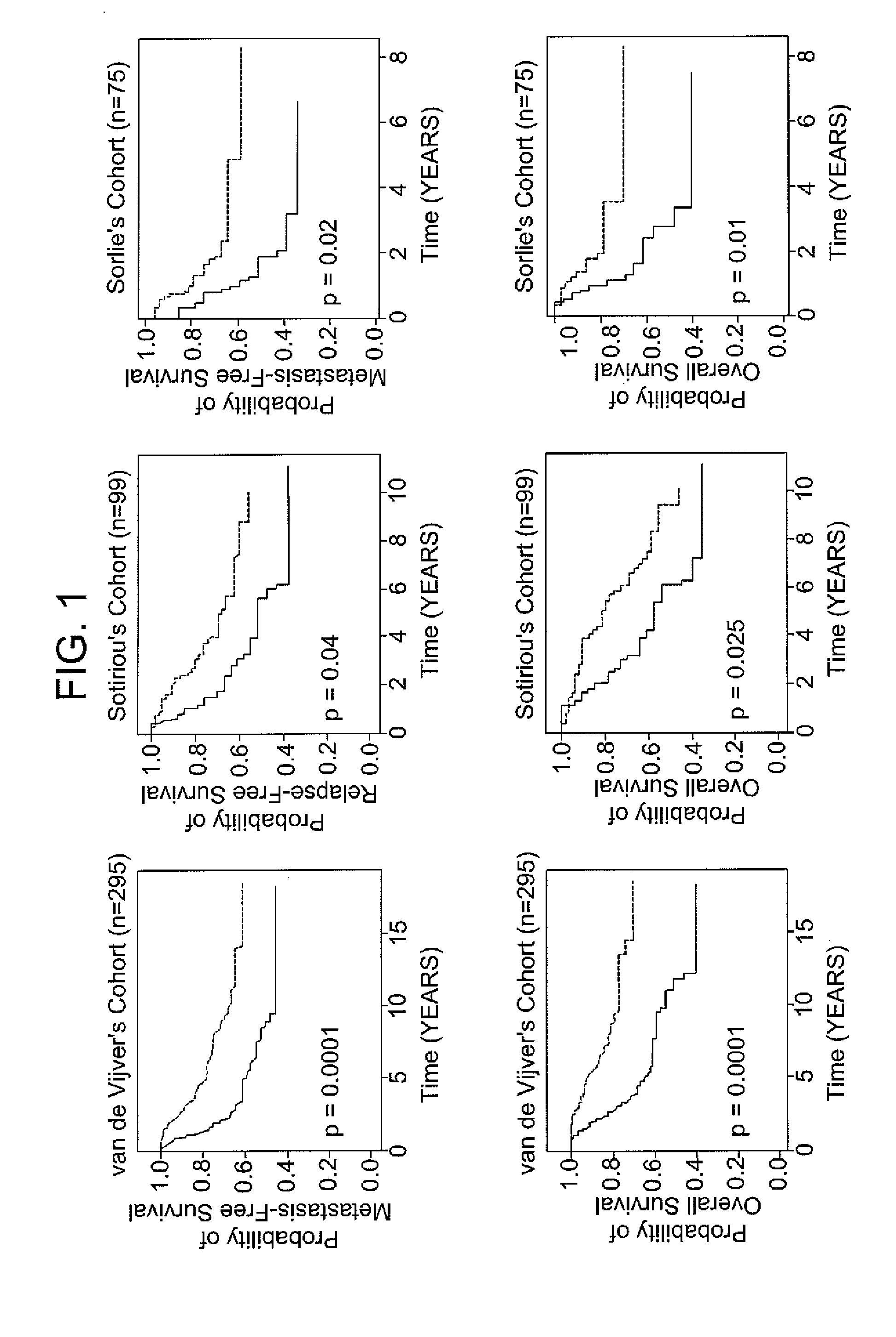
Gene expression signature of genomic instability in breast cancer
InactiveUS20110224089A1Nucleotide librariesMicrobiological testing/measurementNucleic acid sequencingFhit gene
Methods of assessing genomic instability in breast cancer tissue by measuring the expression level of genes CDKN2A, SCYA18, STK15, NXF1, cDNA Dkfzp762M127, p28 KIAA0882, MYB, Human clone 23948, RERG, HNF3A, and ACADSB or a nucleic acid sequence comprising about 90% or greater sequence identity to SEQ ID NO: 21 in breast cancer tissue, an array suitable for use in such methods, and related methods and compositions.
Owner:US DEPT OF HEALTH & HUMAN SERVICES +1
Gene expression signature of genomic instability in breast cancer
Methods of assessing genomic instability in breast cancer tissue by measuring the expression level of genes CDKN2A, SCYA18, STK15, NXF1, cDNA Dkfzp762M127, p28 KIAA0882, MYB, Human clone 23948, RERG, HNF3A, and ACADSB or a nucleic acid sequence comprising about 90% or greater sequence identity to SEQ ID NO: 21 in breast cancer tissue, an array suitable for use in such methods, and related methods and compositions.
Owner:UNITED STATES OF AMERICA +1
Universal polypeptide vaccine and application thereof in preparing drugs for treating/preventing pancreatic cancers
ActiveCN109550044AUniversalVersatilityCancer antigen ingredientsAntineoplastic agentsCancer preventionKRAS
The present invention discloses a universal polypeptide vaccine. The universal polypeptide vaccine comprises at least one of the following 6 mutations: KRAS-G12R, KRAS-G12V, CDKN2A-H83Y, KRAS-Q61H, TP53-R248W and CDKN2A-P94L. Specifically, the universal polypeptide vaccine comprises at least one of the 6 mutations. The universal polypeptide vaccine can be used for preparing drugs for treating / preventing cancers (pancreatic cancers). The universal polypeptide vaccine has relatively good immunotherapeutic effects on the pancreatic cancers.
Owner:HANGZHOU NEOANTIGEN THERAPEUTICS CO LTD
Combination of nelfinavir, metformin and rosuvastatin for treating cancer caused by aberrations in PTEN/TP53
The present disclosure relates to a method of treating cancer using a composition comprising Nelfinavir, Metformin, Rosuvastatin, optionally along with a pharmaceutically acceptable excipient. The said composition is used for the treatment of cancer caused due to aberration in PTEN gene, optionally along with aberration in TP53 gene and related genes selected from group comprising PI3K, CDKN2A, MDM2 and MDM4.
Owner:PRECISION PHARM INC
Combination of mutation sites for detection of type 2 diabetes susceptibility genes, detection primers and applications
ActiveCN107955834BBest detection combinationDetection accuracy mentionsMicrobiological testing/measurementDNA/RNA fragmentationExperimental researchTCF7L2
The invention relates to molecular biology, in particular to a mutation site combination and a primer for detecting susceptible genes of type 2 diabetes as well as an application of the combination and the primer. The mutation site combination comprises 21 mutation sites from KCNQ1, C2CD4A, TCF7L2, SLC30A8, CDKAL1, CDKN2A / 2B, KCNJ11, PPAR2, IGF2BP2, FTO, JAZF1, GRK5, RASGRP1, HHEX, PTPRD and SSR genes. Experimental research finds that the selected mutation site combination can effectively judge whether the sample is type 2 diabetes when used for detecting mutation site genes. The best detection combination is screened out from 21 SNP sites disclosed in the prior art, and the detection accuracy rate is increased to 92% or above on the basis of cost control.
Owner:BEIJING COMPUTING CENT
Method for determining reduced predisposition to cancer based on genetic profile
The invention provide methods for early detection of a reduced risk of developing cancer, which comprises detecting the absence of a series of genetic polymorphisms associated with a predisposition of developing cancer, including the polymorphisms of the genes BRCA1, BRCA2, CARD15 (NOD2), CHEK2, CDKN2A (P16), CYP1B1, FGFR2 (KGFR2), MAP3K1 (MEKK1), p53 (TP53), TNRC9, XPD (ERCC2) and the genetic marker Rs6983267, in a biological sample from the analyzed subject, wherein the absence of the genetic polymorphisms is indicative of significantly decreased risk of developing, at least, breast cancer.
Owner:POMORSKA ACAD MEDYCZNA
Lung cancer targeted drug and chemotherapy drug genomes and application thereof in lung cancer clinical drug treatment
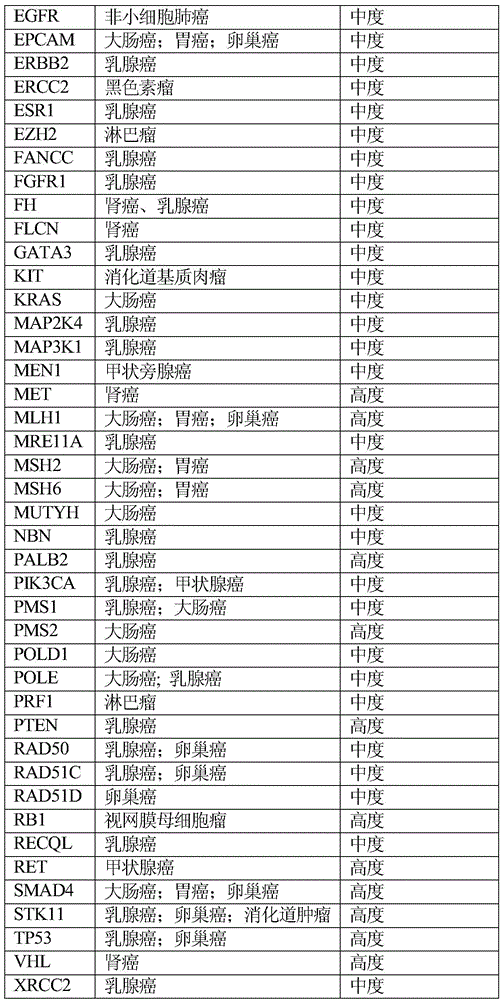
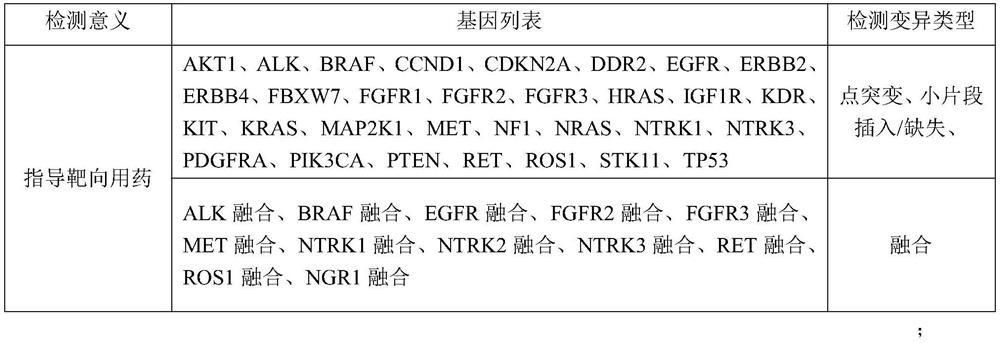
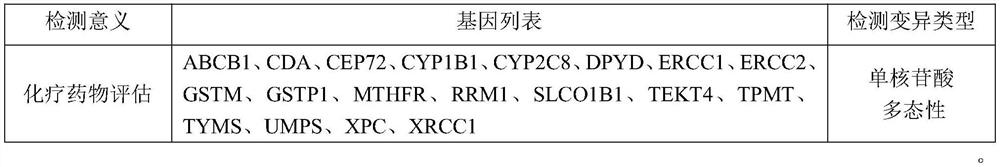
InactiveCN112121170AGood treatment effectSmall toxicityAntineoplastic agentsPharmaceutical active ingredientsSide effectROS1
The invention discloses lung cancer targeted drug and chemotherapy drug genomes and an application thereof in lung cancer clinical drug treatment. The targeted drug genome comprises AKT1, ALK, BRAF, CCND1, CDKN2A, DDR2, EGFR, ERBB2, ERBB4, FBXW7, FGFR1, FGFR2, FGFR3, HRAS, IGF1R, KDR, KIT, KRAS, MAP2K1, MET, NF1, NRAS, NTRK1, NTRK3, PDGFRA, PIK3CA, PTEN, RET, ROS1, STK11 and TP53; and the chemotherapy drug genome comprises ABCB1, CDA, CEP72, CYP1B1, CYP2C8, DPYD, ERCC1, ERCC2, GSTM, GSTP1, MTHFR, RRM1, SLCO1B1, TEKT4, TPMT, TYMS, UMPS, XPC and XRCC1. The lung cancer targeted drug and chemotherapy drug genomes disclosed by the invention are applied to guidance of clinical precise chemotherapy of the lung cancer or auxiliary diagnosis of the lung cancer and curative effect and after-healingmonitoring, can provide the most effective drug selection for lung cancer patients with drug resistance or toxic and side effect intolerance caused by conventional drugs and targeted and chemotherapydrug requirements, improve the drug treatment effect, and reduce the toxic and side effects of the drugs.
Owner:合肥金域医学检验实验室有限公司
Cardiac adenocarcinoma auxiliary diagnostic kit related to a group of genes
The invention relates to a cardia adenocarcinoma auxiliary diagnosis kit related to a group of genes, belongs to the technical field of biomedicine and molecular biology, and particularly relates to an ELISA kit for auxiliary diagnosis of cardia adenocarcinoma. The kit comprises a solid phase carrier and six antibodies which are respectively coated on the solid phase carrier, wherein the six antibodies are as follows: BCL2L1 antibody, DLC1 antibody, CDKN2A antibody, CDKN2B antibody, NRG1 antibody, ErbB4 antibody. Experiments prove that the group of related gene expression product has high expression in the serum of a cardia adenocarcinoma patient, the expression level of the related gene in the human serum is detected by the ELISA kit, and the detection result can be used for auxiliary diagnosis of the cardia adenocarcinoma such as early-onset screening, prognosis evaluation and the like, and the ELISA kit for auxiliary diagnosis of cardia adenocarcinoma has profound clinical significance and important popularization and application prospect.
Owner:THE FIRST AFFILIATED HOSPITAL OF ZHENGZHOU UNIV
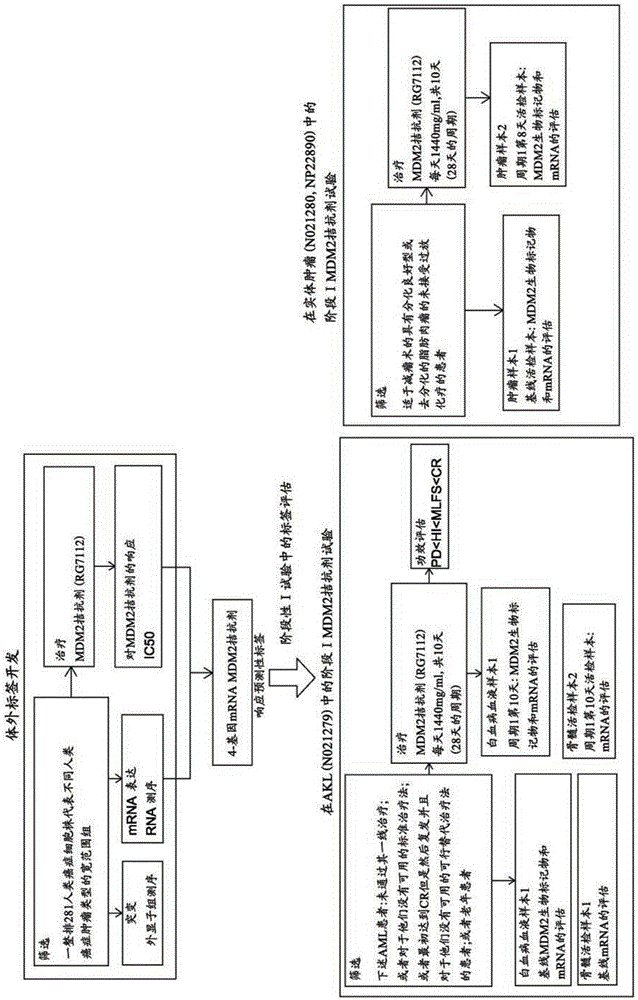
mRNA-based gene expression for personalizing patient cancer therapy with MDM2 antagonist
InactiveCN105378112AMicrobiological testing/measurementAntineoplastic agentsRna expressionCancer therapy
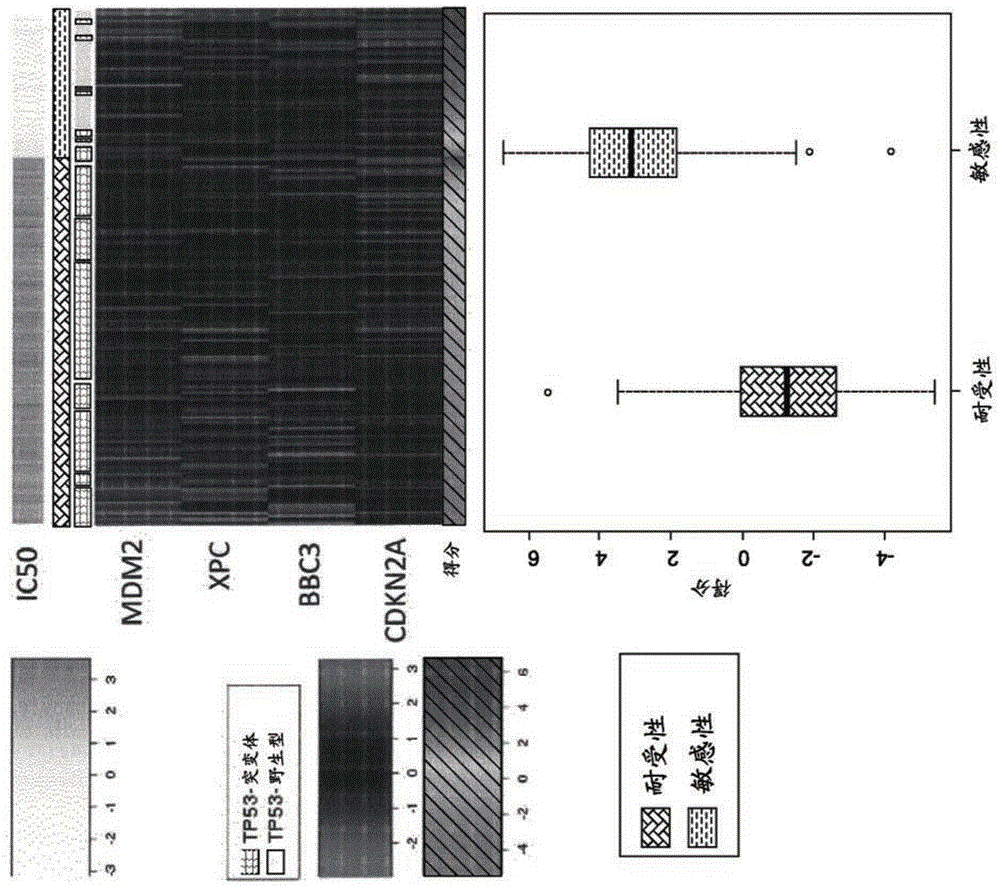
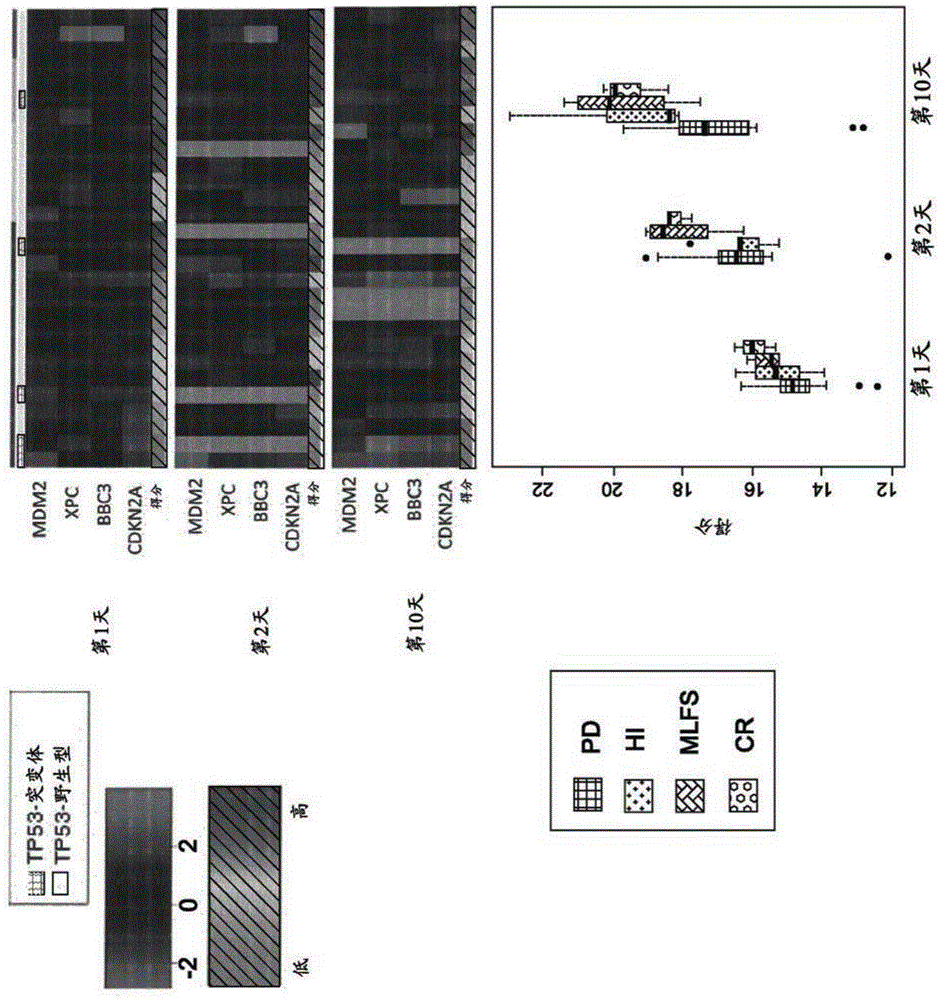
The present application discloses a method to predict responsiveness of a patient, with cancer, to treatment with an MDM2 antagonist of formulae I, II and III as disclosed herein, said method comprising measuring m RNA expression levels of at least MDM2, preferably of a four gene panel comprising MDM2, XPC, BBC3 and CDKN2A, as a biomarker for predicting the response.
Owner:F HOFFMANN LA ROCHE & CO AG
Target genome for gene detection of non-small cell lung cancer patient and related evaluation method, application and kit
ActiveCN112251512AShorten the timeLow costMicrobiological testing/measurementProteomicsOncologyGenome
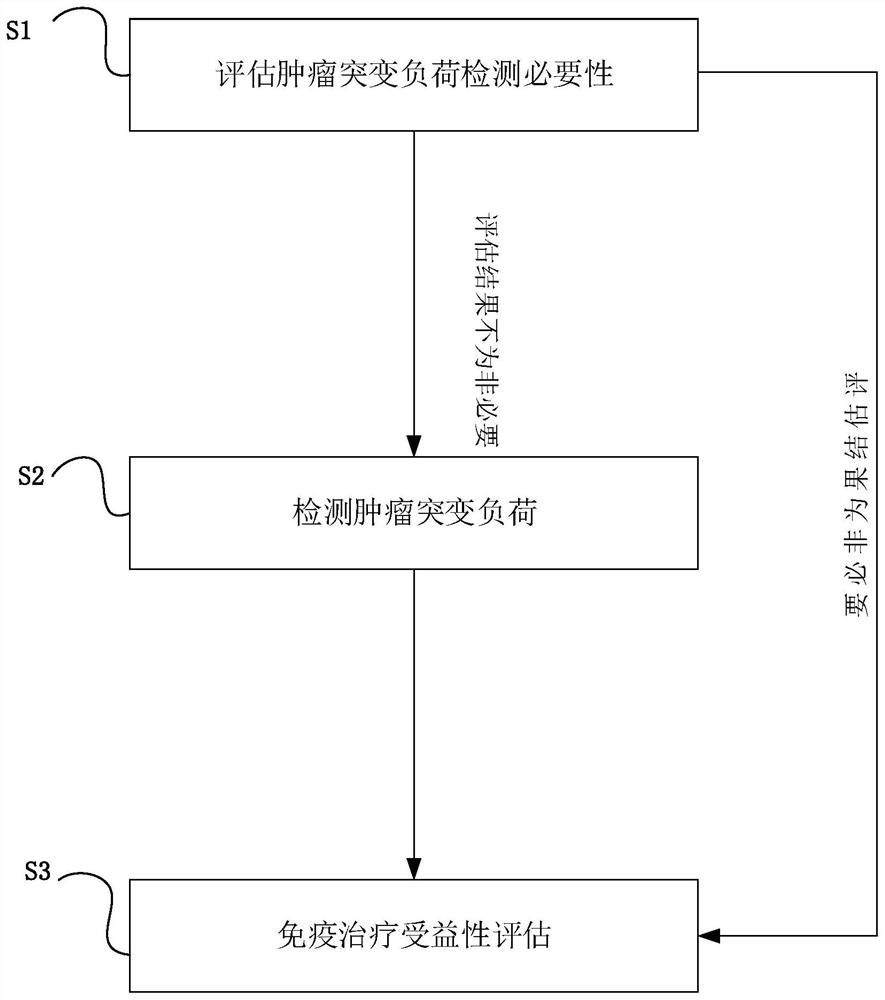
The invention provides a target genome for gene detection of a non-small cell lung cancer patient and a related evaluation method, application and kit. The target genome is used for evaluating whetherthe non-small cell lung cancer patient has the necessity of tumor mutation load detection or not, and the target genome comprises the following genes of EGFR, TP53, CDKN2A and SDHA.
Owner:SHANGHAI ORIGIMED CO LTD
Primer group, kit and method for detecting head and neck tumor gene survival correlation
InactiveCN112708677AAvoid financial burdenImprove guidanceMicrobiological testing/measurementDNA/RNA fragmentationPotential toxicityHead and neck tumors
Owner:AMERICA DIAGNOSIS INC
Universal polypeptide vaccine and its application in the preparation of drugs for treating/preventing pancreatic cancer
ActiveCN109550044BVersatilityShorten the timeCancer antigen ingredientsAntineoplastic agentsCancer preventionPancreas Cancers
The invention discloses a universal polypeptide vaccine, which contains at least one of the following six mutations: KRAS-G12R, KRAS-G12V, CDKN2A-H83Y, KRAS-Q61H, TP53-R248W, CDKN2A-P94L. Specifically: at least one of the polypeptide sequences for the six mutations is included. The universal polypeptide vaccine can be used to prepare medicine for treating / preventing cancer (pancreatic cancer). The universal polypeptide vaccine of the invention has better immunotherapy effect on pancreatic cancer.
Owner:HANGZHOU NEOANTIGEN THERAPEUTICS CO LTD
Marker and model for predicting triple-negative breast cancer chemotherapy response
PendingCN114400069AOptimize treatment planMechanical/radiation/invasive therapiesProteomicsPRC1Oncology
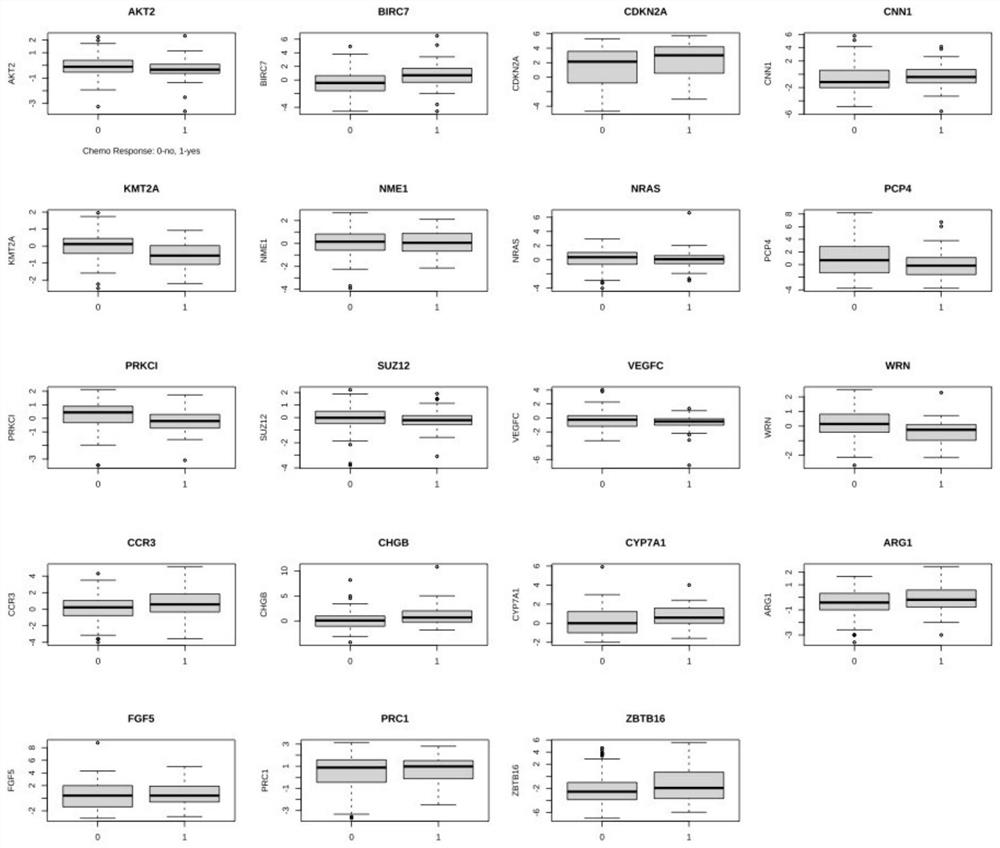
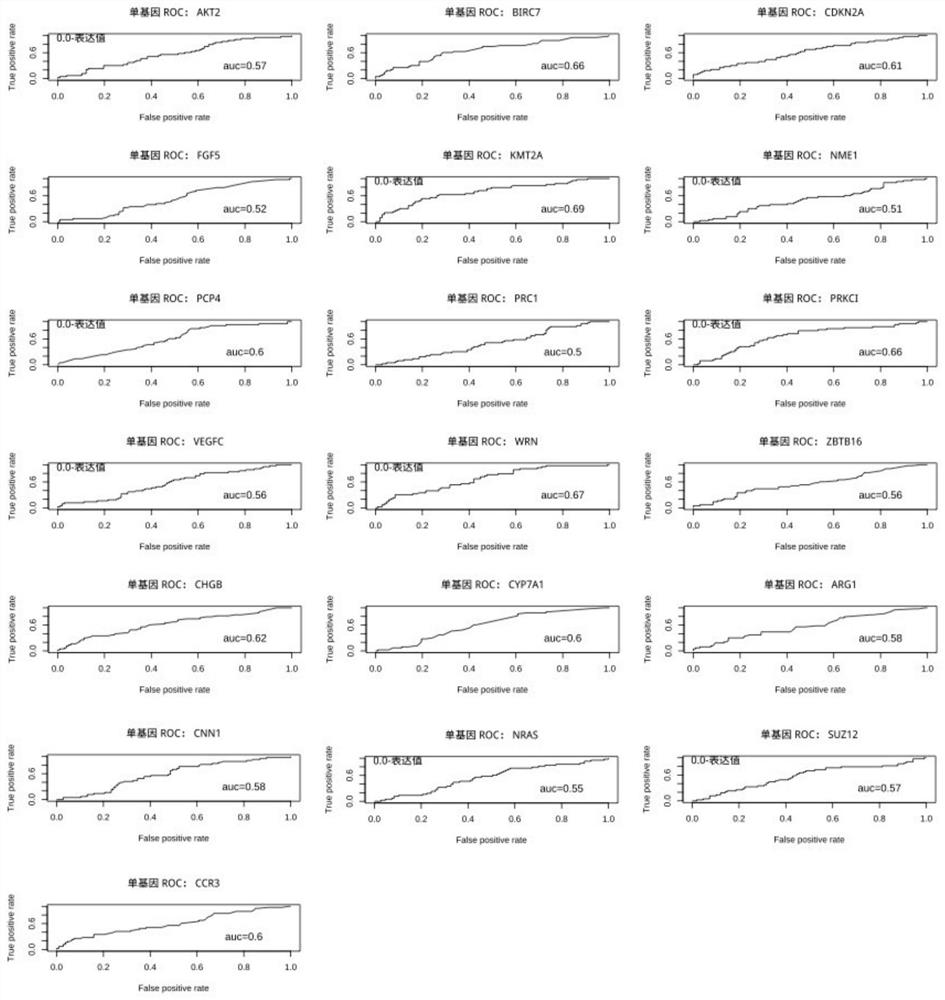
The invention relates to a marker and a model for predicting chemotherapy response of triple negative breast cancer. The marker comprises one or a combination of more than two of the following genes: AKT2, ARG1, BIRC7, CCR3, CDKN2A, CHGB, CNN1, CYP7A1, FGF5, KMT2A, NME1, NRAS, PCP4, PRC1, PRKCI, SUZ12, VEGFC, WRN and ZBTB16. According to the method, the chemotherapy response is taken as a target function, and the accurate model for predicting the chemotherapy response of the triple-negative breast cancer is established by utilizing the whole transcriptome data of the triple-negative breast cancer slice based on an iterative linear regression algorithm of correlation coefficient layering. By utilizing the model, evaluation can be performed before chemotherapy of a triple-negative breast cancer patient, and the model is used for guiding establishment of a treatment scheme.
Owner:SHENZHEN LUWEI BIOTECHNOLOGY (BIOMANIFOLD TECH CO) LTD
A detection of cervical cancer marker cdkn2a antigen epitope polypeptide and application
ActiveCN104558147BHigh expressionGuide early diagnosisTumor rejection antigen precursorsTumor specific antigensSerum igeEpitope
The invention relates to a CDKN2A epitope peptide for detecting cervical cancer markers and an application of the CDKN2A epitope peptide, and belongs to the technical field of biology. An amino acid sequence of the CDKN2A epitope peptide for detecting the cervical cancer markers is as shown in SEQ ID NO:1; and the CDKN2A epitope peptide for detecting the cervical cancer markers is applied to preparation of an early diagnosis kit for cervical cancer. The CDKN2A epitope peptide has the advantages that the autoantibody expression level in serum and plasma of a patient with the cervical cancer is detected and a corresponding reagent is developed; the dangerousness of the cervical cancer is predicted; and a foundation is laid for preparation of the early diagnosis kit for the cervical cancer.
Owner:刘林林
Features
- R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
Why Patsnap Eureka
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Social media
Patsnap Eureka Blog
Learn More Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com