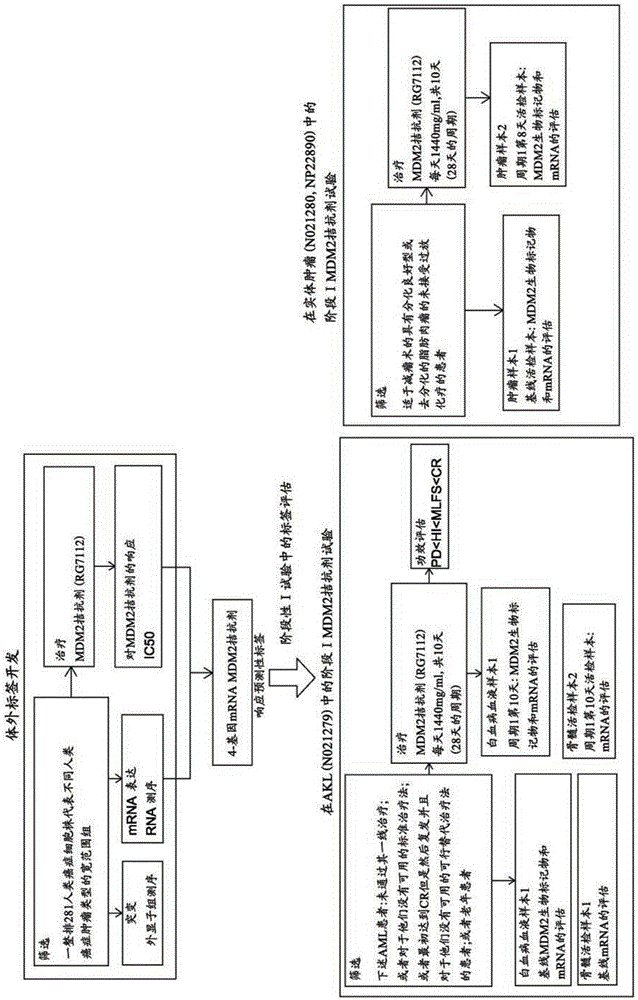
mRNA-based gene expression for personalizing patient cancer therapy with MDM2 antagonist
An MDM2, patient technology, applied in the direction of antitumor drugs, microbial determination/test, biochemical equipment and methods, etc., can solve the problems of unmet clinical development of MDM2 antagonists and MDM2 antagonists.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0271] RNA sequencing of CELLO cell lines
[0272] RNA sequencing (RNA-seq) by next-generation sequencing (NGS) technology is a precise and sensitive method for measuring gene expression, where additional kinetics are used to detect alternative splicing, allele-specific expression, non-coding RNA, and Various forms of mutations (SNPs, insertions and deletions, gene fusions). The NGSIlluminaHiSeq machine generates raw base calls as reads of 50 or 100 bp length, which are subjected to several data analysis steps. RNA-seq is performed at 40 to 50 million reads / sample. This number provides relatively high sensitivity for detection of low expressed genes while allowing multiplexing of samples that can be scaled. RNA was prepared from standard kits, while RNA libraries were prepared from polyATruSeq Illumina kits. For each RNA-seq reaction, 100 ng of mRNA / cell line was used. For each specimen, multiple quality control procedures were applied to the RNA-seq data. The Illumina Hi...
Embodiment 2
[0274] Exom Sequencing of CELLO Cell Lines
[0275] Compound A in vitro test
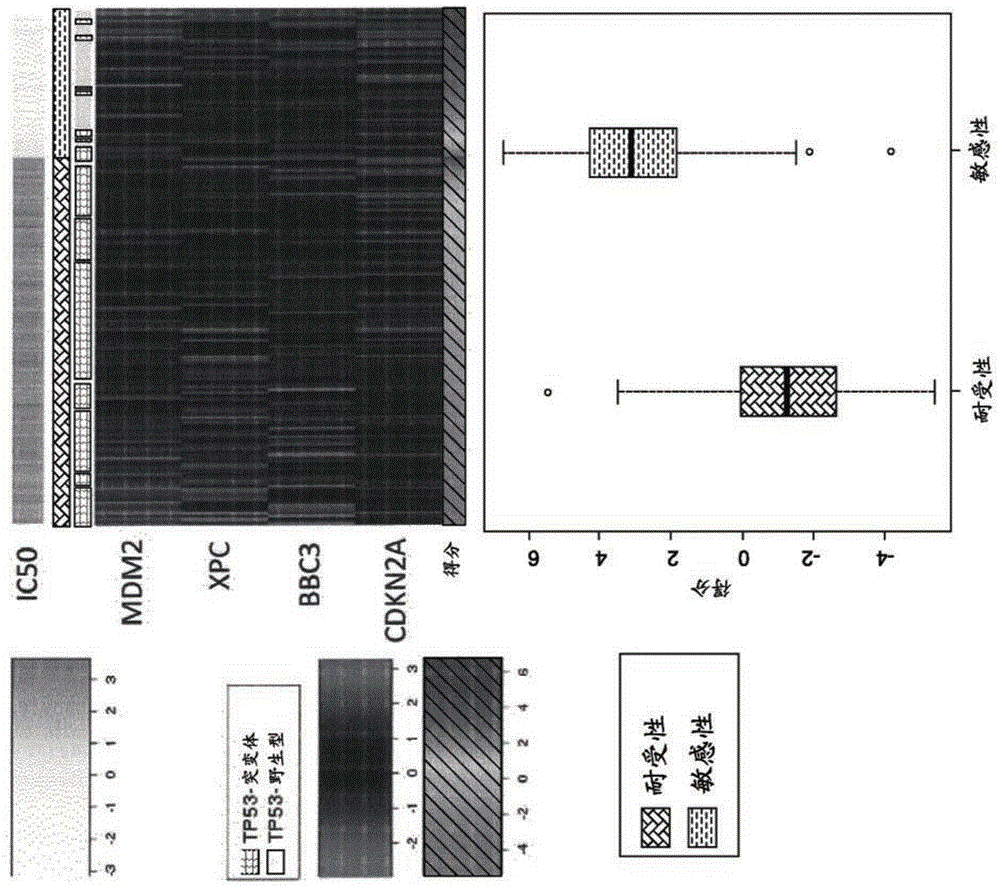
[0276] Treatment of 281 cells with MDM2 antagonists resulted in IC50 (therapeutic response). Cell lines are derived across a wide range of tumor cell types.
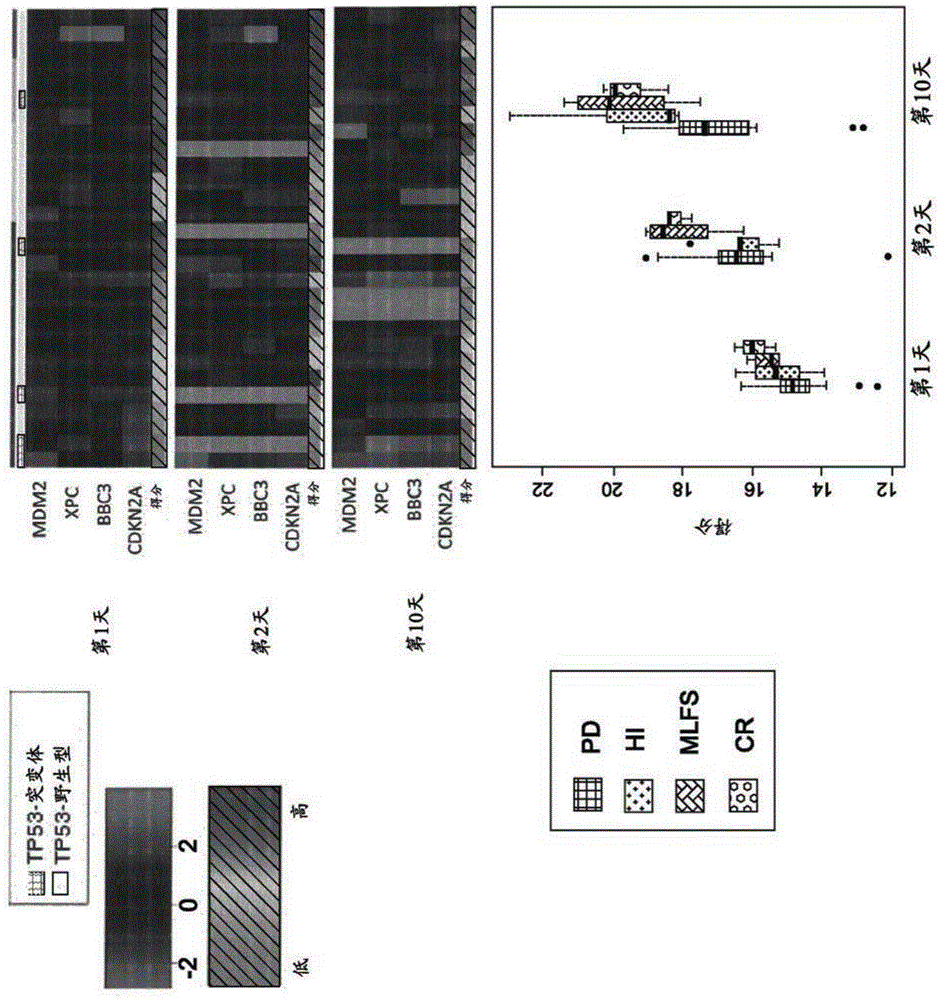
[0277] After careful annotation to remove low-quality cells and germline mutations, of the 281 cell lines evaluated, 210 cell lines showed mutations in TP53. Of the 7 AML cell lines tested, 5 showed mutations in TP53. Cell lines harboring mutant TP53 were far less sensitive to MDM2 antagonist treatment (P-16 ), which is consistent with previously published data. Genome-wide association between baseline mRNA expression and MDM2 antagonist treatment response (IC50) identified a list of 13 genes that were significantly associated with a P-value of 2.38x10 -47 to 9.56x10 -23 (Supplementary Table 2). Functional annotation indicated that 13 important genes from genome-wide association analysis with correlation coefficients ranging from -0.47 t...
Embodiment 3
[0288] In vitro test gene expression analysis method
[0289] Messenger RNA (mRNA) expression levels at baseline were measured by RNA sequencing (RNA-seq) and microarray using GeneChipHumanGenomeU133Plus2.0Array before MDM2 antagonist treatment. Gene expression analysis of RNA-seq data is summarized here. First, sequence reads were matched to the reference human genome and to other databases of splice junction fragments derived from known exon positions on the reference genome (Cufflinks software). These matching reads are then combined to produce discrete counts of reads (or bases that determine the sequence) per gene. These gene expression counts were then normalized to equalize the total amount of RNA counts per sample and corrected for gene / transcript length (RPKM). All genes with expression values less than RPKM = 1 in all cell lines were removed (this roughly equates to an expression level of less than one copy of RNA molecule / cell). Next, the normalized gene counts...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com