Computer coding method for double-stranded nucleotides of DNA coding compound library
A coding method and nucleotide single-strand technology, applied in chemical libraries, calculations, combinatorial chemistry, etc., can solve the problem of not considering differences, limiting the number of small molecules, not considering the impact of fragment compound reaction success or not, etc. question
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
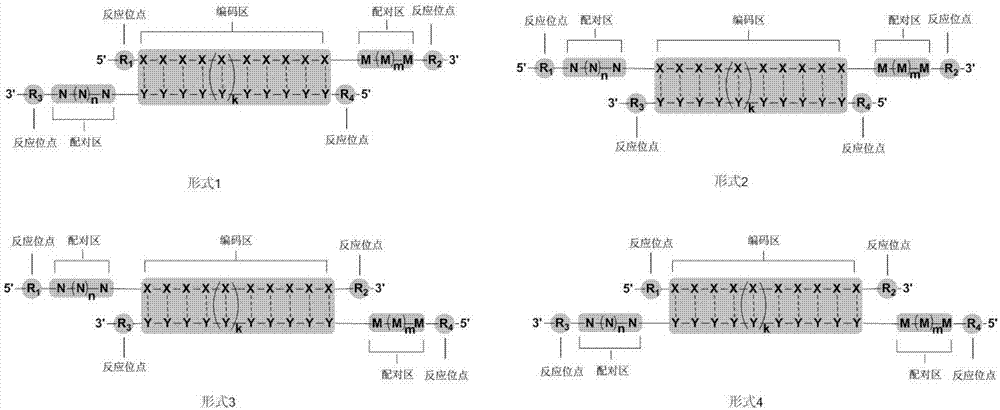
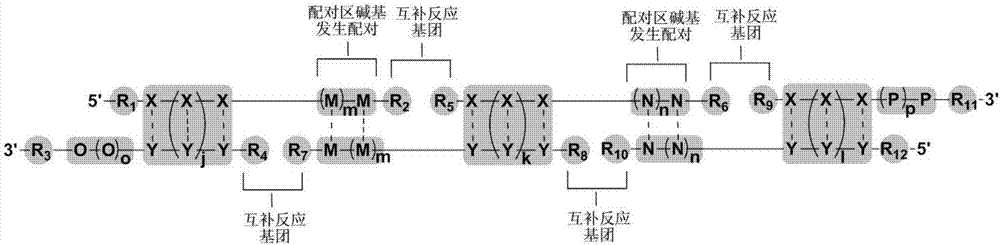
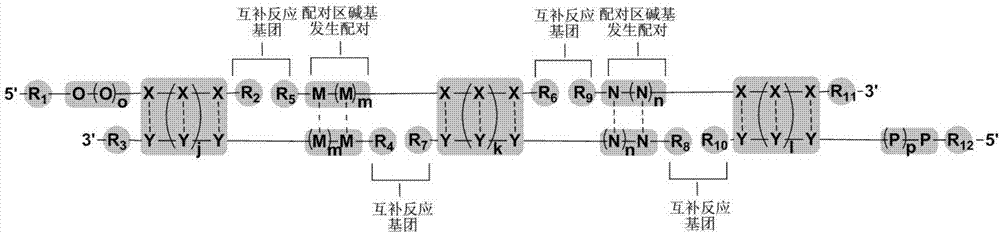
Image
Examples
Embodiment 1
[0087] Example 1, the computer coding method for a nucleotide double-strand with a Hamming distance of 9 bp and a Hamming distance greater than or equal to 4 in the coding region at the 3' end of the single-strand with both protruding base sequences
[0088] 1. Computer coding method for nucleotide double strands with 9 bp coding region and Hamming distance greater than or equal to 4
[0089] This encoding uses software shell and Python language, but if the base sequence of the specific nucleotide double-strand obtained by using a similar encoding method is completely identical to the base sequence of the present invention by using other similar software, it should be understood as Within the scope of patent protection of the present invention.
[0090] The specific code is as follows:
[0091]
[0092]
[0093]
[0094] A total of 632 pairs of specific base sequences were obtained in the nucleotide double strands with a Hamming distance greater than or equal to 4. ...
Embodiment 2
[0136] Example 2, two 6-base overhangs are all at the 3' end of the single strand, the coding region is 6bp and the Hamming distance is greater than or equal to 4 nucleotide double-stranded computer coding method
[0137] 1. Computer coding method for nucleotide duplexes with a coding region of 6 bp and a Hamming distance greater than or equal to 4
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com