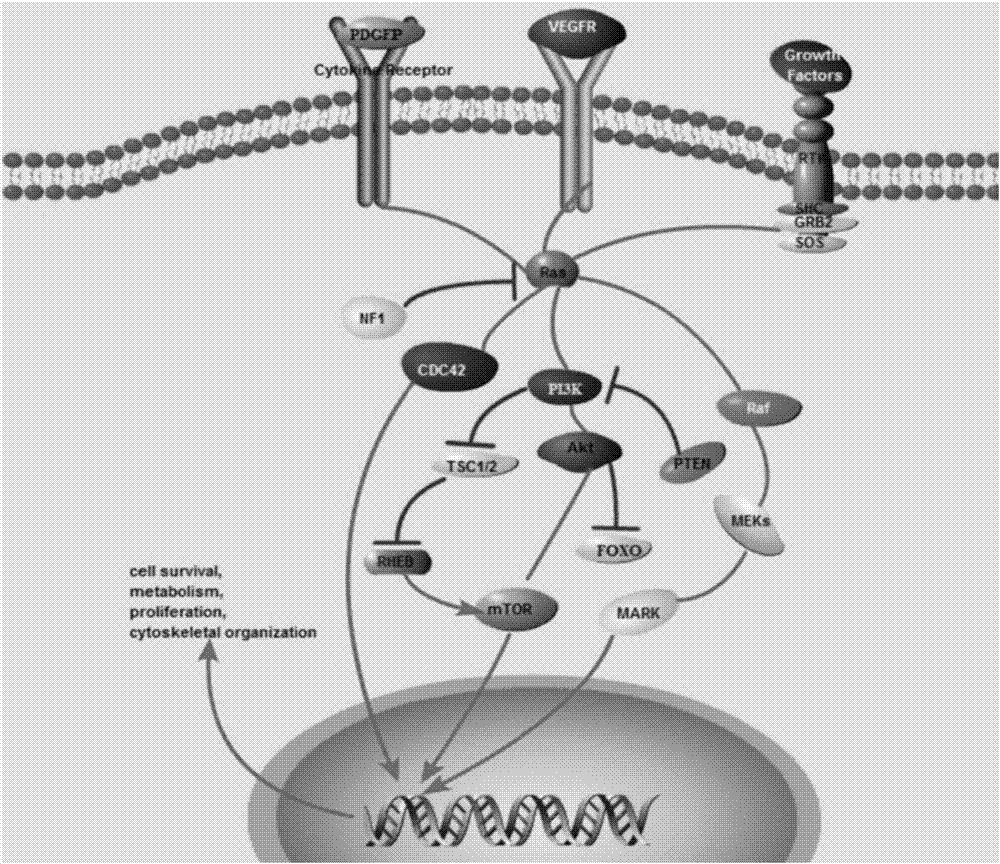
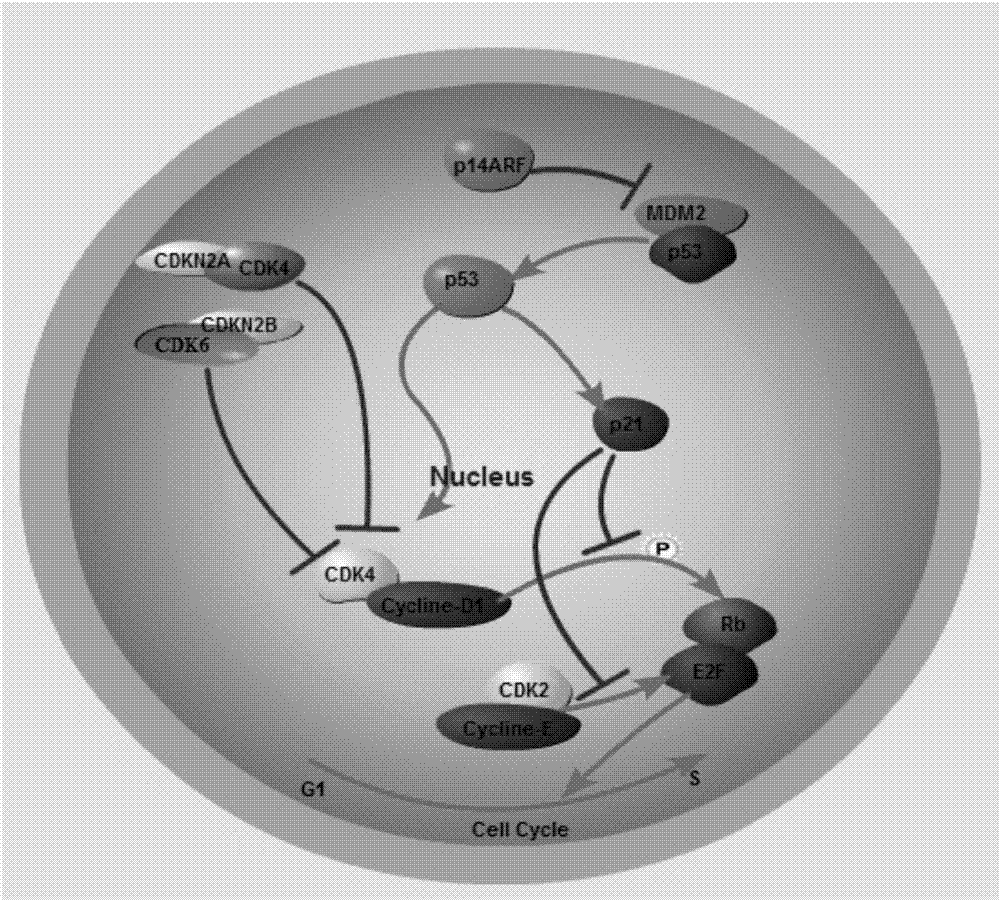
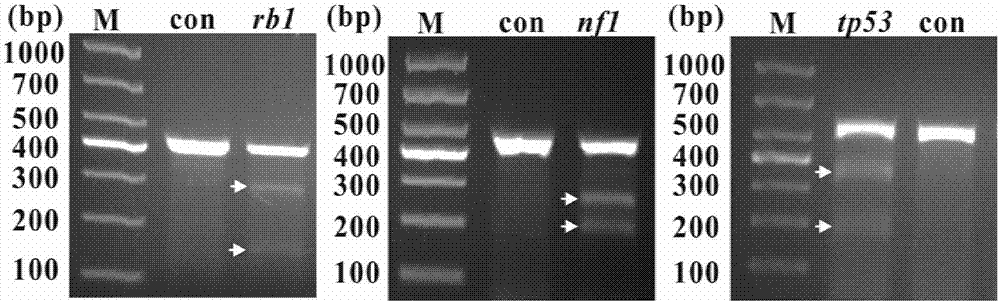
Gene knockout vector and zebra fish glioma model thereof
A gene knockout and zebrafish technology, applied in the direction of using vectors to introduce foreign genetic material, genetic engineering, other methods of inserting foreign genetic material, etc., can solve the problems of animal embryo lethality, difficulty in applying the pathogenesis of glioma patients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Establishment of zebrafish glioma model
[0079] Step 1: Construct the gRNA sequence of the targeting sequence
[0080] Firstly, the CDs sequences of the rb1nf1 and tp53 genes of zebrafish were found on the website of NCBI (National Center for Biotechnology Information, USA). Find the target site sequence on the chopchop website, the target sequences of the three locked genes are
[0081] rb1:TGCATGGAGAATATGGGAGA;
[0082] nf1:GGCGCACAAGCCCGTGGAAT;
[0083] tp53:TGATTGTGAGGATGGGCCTG.
[0084] Then, to construct the targeting sequence gRNA, synthesize the following primers:
[0085] rb1-gRNA-F: TAATACGACTCACTATAGGGTGCATGGAGAATATGGGAGAGTTTTTAGAGCTAGAAATAGC
[0086] nf1-gRNA-F: TAATACGACTCACTATAGGGGCGCACAAGCCCGTGGAATGTTTTTAGAGCTAGAAATAGC
[0087] tp53-gRNA-F: TAATACGACTCACTATAGGGTGATTGTGAGGATGGGCCTGGTTTTAGAGCTAGAAATAGC
[0088] gRNA-R:AGCACCGACTCGGTGCCAC
[0089] Among them, gRNA-R is a general downstream primer synthesized by three gRNAs.
[0090]Using the plasmi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com