Group of HLA-related SNP marks and detection primer and determination method thereof
A certain method and marker technology, applied in the medical field, to achieve the effect of assisting the assessment of transplant rejection risk and autoimmune disease
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
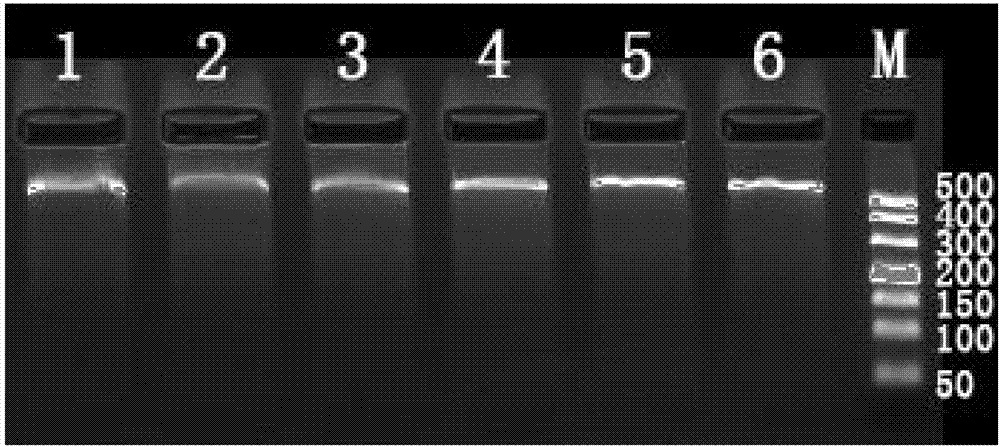
[0038] Embodiment 1 Obtaining a group of HLA-associated SNP markers
[0039] 1.1 Obtaining of blood samples in Example 1
[0040] Example 1 The blood samples were 6 blood samples randomly drawn from the pre-transplant whole blood samples of kidney transplant patients collected by the Organ Transplantation Department of the First Affiliated Hospital of Sun Yat-sen University for genomic DNA extraction.
[0041] 1.2 Example 1 Extraction of Genomic DNA from Blood Sample
[0042]In this experiment, TIANamp Blood DNAKit (catalogue number: DP348) was used to extract genomic DNA from the blood sample of Example 1. The specific steps are as follows:
[0043] (1) Gently invert and mix the whole blood samples of the six patients in Example 1, and draw 200ul from it into a new 1.5ml EP tube (if the DNA extraction concentration is too low, you can also centrifuge the blood sample at 500g for 5min to separate the layers, and take The buffy coat cells in the middle are 200ul for subsequen...
Embodiment 2
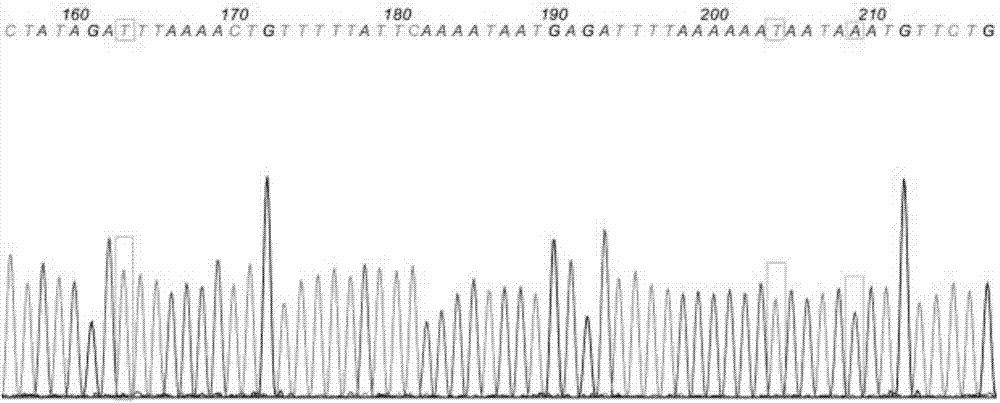
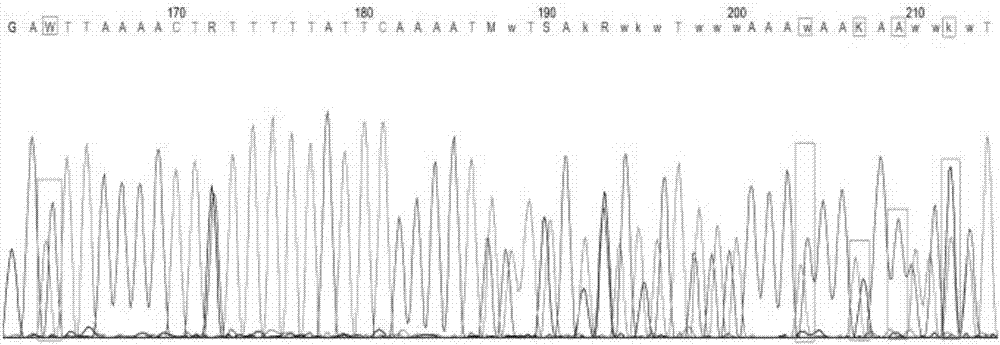
[0077] The difference between this embodiment and the embodiment is that the primers used in the amplification have the nucleotide sequences shown in SEQ ID NO: 1 and SEQ ID NO: 5; forward sequencing can be used during sequencing, and reverse sequencing can also be used. For sequencing, the forward sequencing primer is SEQ ID NO:1, and the reverse sequencing primer is SEQ ID NO:5.
Embodiment 3
[0079] The difference between this embodiment and the embodiment is that the primers used in the amplification are the nucleotide sequences shown in SEQ ID NO:3 and SEQ ID NO:5; forward sequencing can be used during sequencing, and reverse sequencing can also be used , during sequencing, the forward sequencing primer is SEQ ID NO:3, and the reverse sequencing primer is SEQ ID NO:5.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com