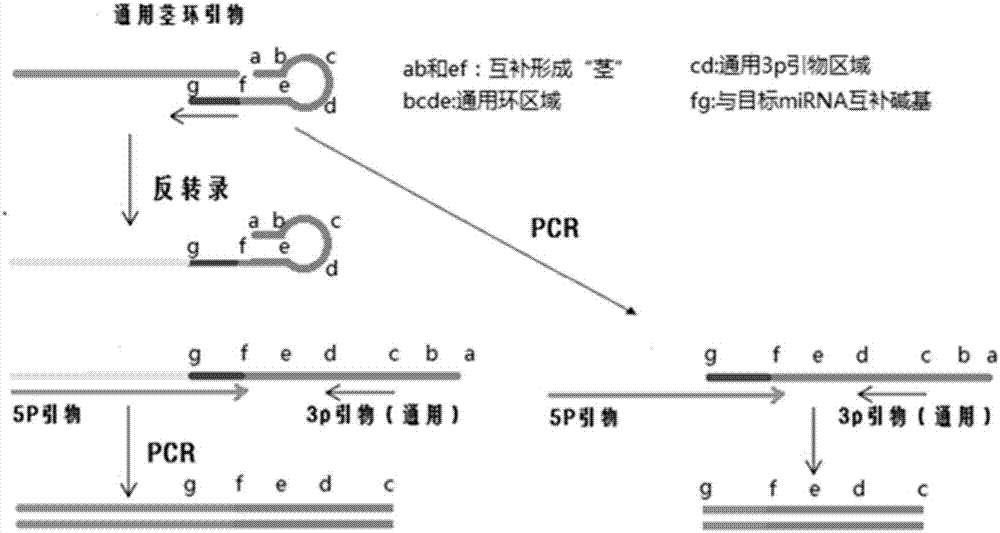
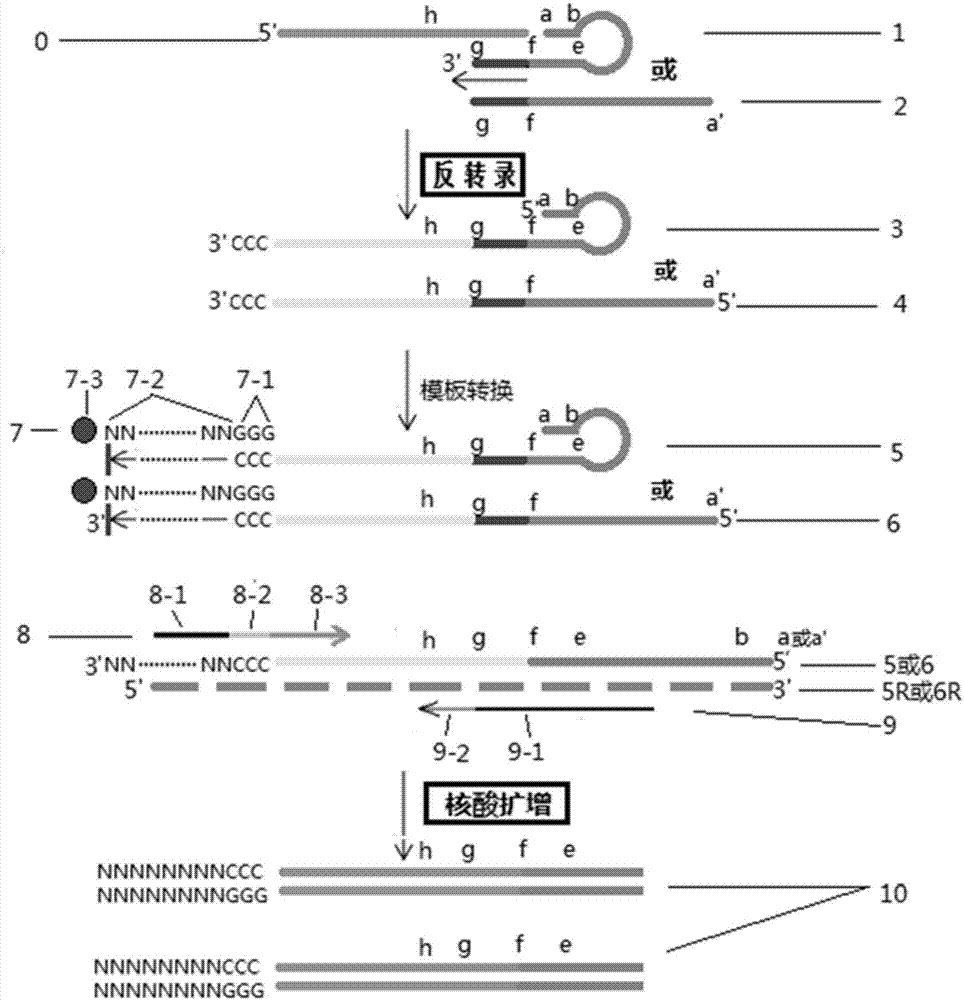
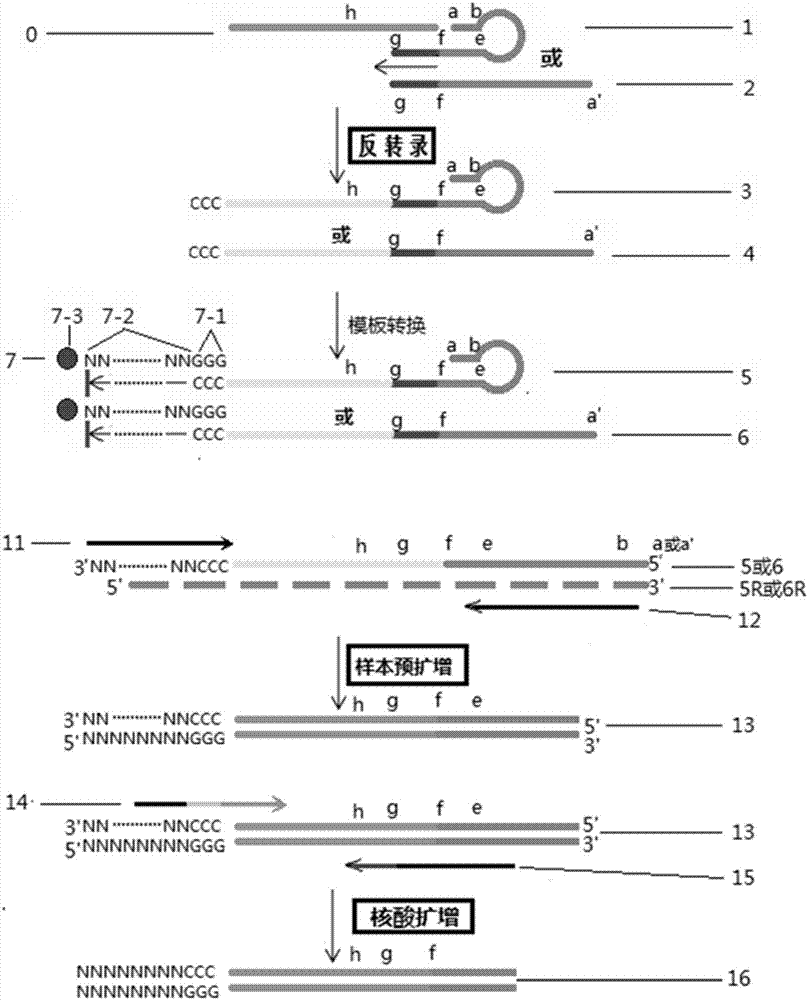
Short fragment nucleic acid chain detection method and pre-amplification method
A detection method and pre-amplification technology, applied in biochemical equipment and methods, microbial determination/inspection, etc., can solve problems such as difficult binding, primers cannot work, and Tm value is increased to an appropriate temperature.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0085] Example 1 Quantitative detection of miRNA - real-time fluorescent PCR method
[0086] The purpose of the experiment: to compare the background amplification of the miRNA quantitative detection method designed according to the idea of the present invention and the general 3' stem-loop primer method
[0087] Experimental design ideas: select 3 miRNAs to design universal stem-loop primers (4 bases at the end are different, respectively bind to the target miRNA, and the stem-loop structure is common), and design 4 sets of experiments, namely:
[0088] Group A: universal 3' primer experimental group, using RNA as template, reverse transcription primer: stem-loop primer, PCR primer: 5' specific primer, 3' universal primer;
[0089] Group B: Universal 3' primer control group, no RNA template, and PCR with group A PCR primers;
[0090] Group C: the experimental group of the present invention, using RNA as a template, reverse transcription primer: stem-loop primer, adding TSO...
Embodiment 2
[0104] Example 2 Quantitative detection of miRNA - real-time fluorescent PCR method
[0105] The purpose of the experiment: to compare the amplification efficiency of the miRNA quantitative detection method designed according to the idea of the present invention and the semi-nested PCR method
[0106] Experiment design ideas: Design 2 groups of experiments for 8 miRNAs, namely:
[0107] Group E: Reverse transcription primers: special stem-loop primers; PCR primers: 5’ and 3’ specific primers designed according to the principles of background literature 2
[0108] Group F: reverse transcription primers: special stem-loop primers+TSO sequence; PCR primers: 5' and 3' specific primers designed according to the design principles of the present invention
[0109] The specific miRNA, reverse transcription primer sequence, TSO sequence, and cDNA amplification primer sequence are numbered in the sequence table as follows:
[0110]
[0111] Total RNA: artificially synthesized abo...
Embodiment 3
[0124] Example 3 Pre-amplification of low-abundance samples
[0125] Experimental purpose: to test the sensitivity / detection rate and specificity / specificity of the sample pre-amplification method designed by the idea of the present invention
[0126] Experiment design idea: low-abundance samples were used, divided into pre-amplification group and non-pre-amplification group, and the expression quantitative results of the three miRNAs in Example 1 were tested.
[0127] Group G: No pre-amplification group. After reverse transcription, the pre-amplification components were added together, and placed at 4°C until the pre-amplification of group B was completed, and PCR amplification was performed simultaneously with group B.
[0128] Group H: pre-amplification group.
[0129] The reverse transcription primers, TSO sequence, and PCR primers all used the primers of Group C of Example 1, and the preamplification primer sequences: 5' primer SEQ No.71, 3' primer SEQ ID No.8.
[013...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com