Colorectal cancer early screening primer group based on four genes and kit
A colorectal cancer, primer set technology, applied in the biological field, can solve the problems of high false negatives in detection, affecting the judgment of results, etc., to improve the accuracy and comprehensiveness, reduce the proportion of false negatives, and reduce the proportion of false negatives. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1 Detection of primer sequences for exon mutations in four genes of APC, CTNNB1, B-raf and K-ras
[0037] (1) A group of primers for detecting mutation sites of all exons of APC, CTNNB1, B-raf, and K-ras genes
[0038]A group of primers for the whole exon mutation sites of APC, CTNNB1, B-raf, and K-ras genes are as follows:
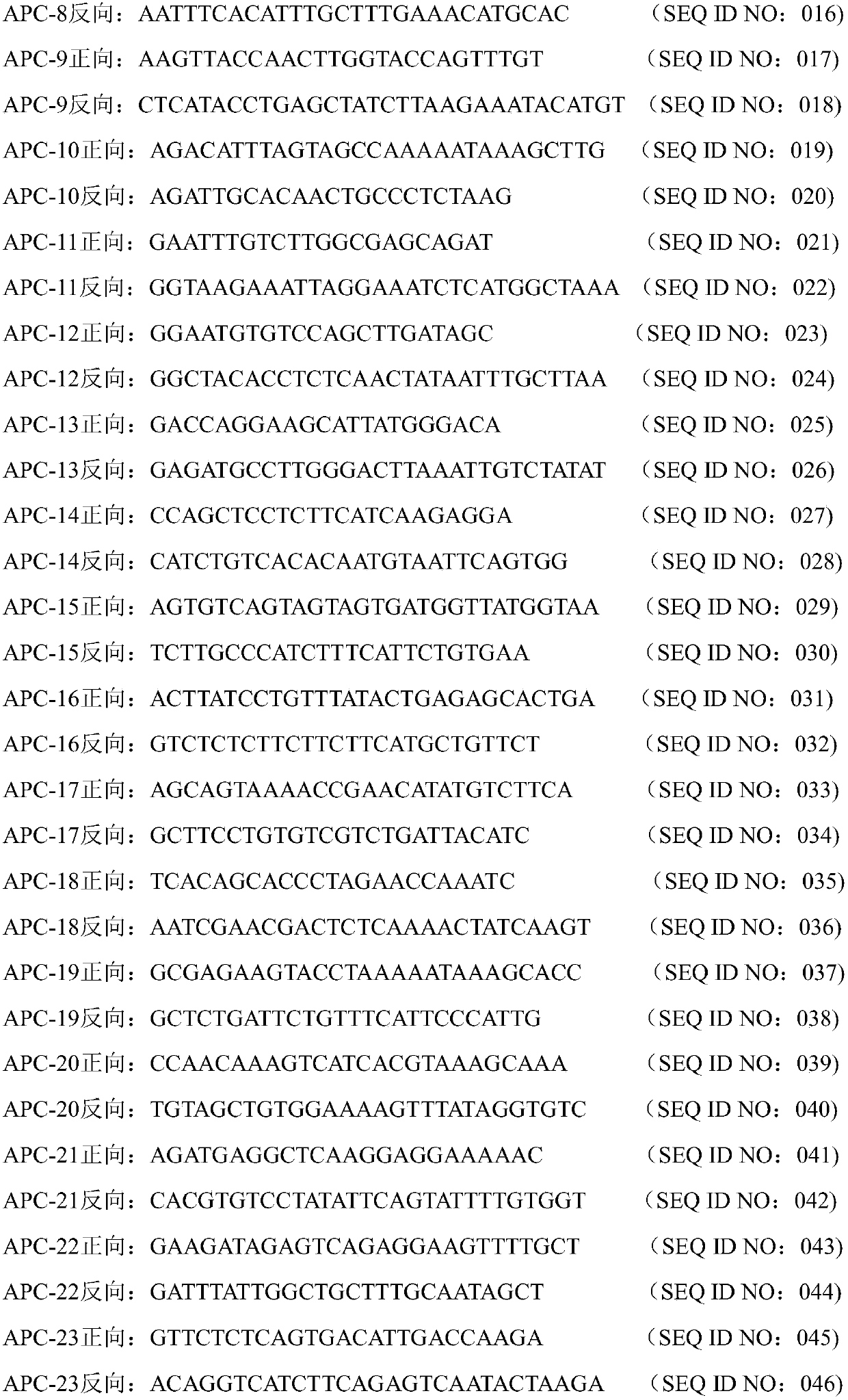
[0039] The sequence shown in SEQ ID NO: 001-058 is the forward and reverse primers for amplifying and covering the whole exon mutation site of APC gene;
[0040] The sequence shown in SEQ ID NO: 117-136 is the forward and reverse primers for amplifying the mutation site covering the whole exon of CTNNB1 gene;
[0041] The sequence shown in SEQ ID NO: 157-184 is the forward and reverse primers for amplifying and covering the whole exon mutation site of B-raf gene;
[0042] The sequences shown in SEQ ID NO: 211-216 are forward and reverse primers for amplifying and covering the whole exon mutation site of K-ras gene.
[0043] In the detect...
Embodiment 2
[0051] Example 2 The method for detecting hotspot mutations in the whole exons of four genes APC, CTNNB1, B-raf and K-ras
[0052] (1) Genomic DNA extraction from feces
[0053] 1) Take 200mg of feces in a test tube (if the stool sample is liquid, take 200μL in the test tube), add 2mL of PBS to shake and mix well, centrifuge at 3000g for 5 minutes, and collect the supernatant; take 1mL of the collected supernatant in 1.5mL In a centrifuge tube, centrifuge at 12,000 rpm for 5 minutes, discard the supernatant, and resuspend the pellet with 1 mL of PBS. After shaking and mixing, centrifuge at 12,000 rpm for 5 minutes to collect the pellet.
[0054] 2) Add 200 μL of 1% SDS, suspend the precipitate, and place it at 37° C. for 30 minutes.
[0055] 3) Add 200 μL of proteinase K and 20 μL of digestion solution, shake and mix well, and bathe in water at 56°C for 10 minutes.
[0056] 4) Add 600 μL of 15% ethanol, gently invert and mix well. If there is a translucent suspension, it wil...
Embodiment 3
[0082] Example 3 Using the primers and methods provided by the present invention to detect clinical samples
[0083] Six clinical stool samples were taken from the hospital, all of which were not determined to be colorectal cancer patients, and whether there were four gene exon mutations of APC, CTNNB1, B-raf and K-ras in the 6 samples. Genomic DNA was extracted according to the method described in Example 2, and reagents were prepared and detected. After each sample was sequenced, the 55 known hotspot mutations were compared with the wild-type genome sequence, and finally based on the sequencing results, it was preliminarily judged whether the patient had new mutations in the four genes and whether he was a colorectal cancer patient.
[0084] The test results are shown in the table below:
[0085] Sample serial number
Sequencing results
colorectal cancer patients
1
no mutation
no
2
no mutation
no
3
K-ras G12D
yes
...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com