Detection primer group for Paecilomyces hepiali Chen et Dai in ophiocordyceps sinensis culture solution, detection kit and detection method
A technology of Paecilomyces bat moth and Cordyceps sinensis, which is applied in the field of bioengineering, can solve the problems of restricting the development of Cordyceps sinensis artificial cultivation industry, the quality of bacterial liquid is difficult to guarantee, and the sampling microscope inspection misses detection, etc., and achieves low cost, simple method, and primer highly specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] 1. The detection primer set of Paecilomyces spp. in the culture medium of Cordyceps sinensis
[0032] The detection primer set includes 2 pairs of specific primers for Paecilomyces hematoptera:
[0033] The first pair of specific primers:
[0034]Ph816F: 5'-CCTTTTGTGAACATACCTATC-3' (such as figure 1 Shown) (as shown in SEQ ID NO.1);
[0035] Ph1302R: 5'-GAGGTCAACGTTCAGAAGTC-3' (such as figure 1 Shown) (as shown in SEQ ID NO.2);
[0036] The second pair of specific primers:
[0037] Ph935F: 5'-GTATCTTCTGAATCCGCCGCA-3' (eg figure 1 Shown) (as shown in SEQ ID NO.3);
[0038] Ph1162R: 5'-CCGATTTCCCCAAAGGGAAG-3' (such as figure 1 Shown) (as shown in SEQ ID NO.4).
[0039] 2. PCR template preparation
[0040] Take the purified cultured Cordyceps sinensis Ophiocordyceps sinensis (Berk) mycelium and Paecilomyces hepiali Chen et Dai mycelia 100mg each in 2mL Microfuge tubes, grind, respectively according to the fungal CTAB genome extraction method and Fungal Genomic DN...
Embodiment 2
[0062] Embodiment 2: sensitivity analysis
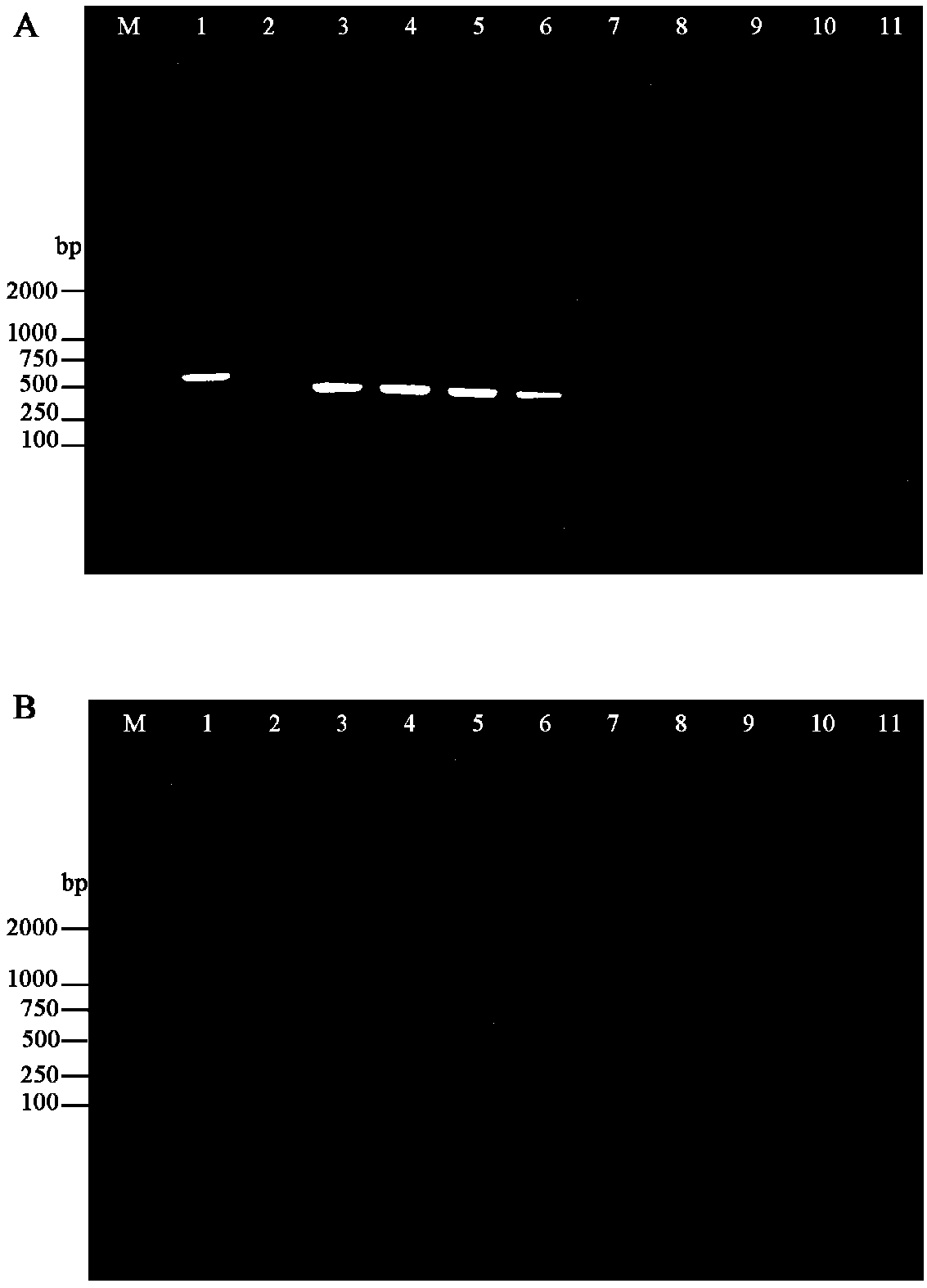
[0063] Genomic DNA of purified Cordyceps sinensis and Paecilomyces hematalis was used to detect the concentration and quality of genomic DNA samples with a micro-nucleic acid spectrophotometer and electrophoresis. Then prepare 10 PCR templates according to the amount of genomic DNA of two kinds of bacteria, see Table 1, carry out nested PCR amplification (with embodiment 1 step 3) respectively with template 1~10 as template, agarose gel electrophoresis detects PCR product According to the band size, the sensitivity and accuracy of the detection primer set and detection method were confirmed.
[0064] The electrophoresis detection result of the PCR reaction product of the first round of PCR amplification shows that when the PCR template DNA (30ng of the total amount of Genomic DNA of Cordyceps sinensis and Paecilomyces thaliana) only accounts for 10 -5 (i.e. 0.0003ng), the target band of the reaction product was clear, and when the a...
Embodiment 3
[0068] Example 3: Application Analysis
[0069] In this example, 16 bottles of Cordyceps sinensis culture solution were randomly selected, and 1 mL of bacterial solution was taken from each bottle of culture solution into a 2 mL centrifuge tube, and sterile water was added, centrifuged at 8000 rpm at 10°C for 15 min, the eluate was discarded, and the sample was quickly placed in Quick-freeze in liquid nitrogen, grind, extract genomic DNA with reference to the instructions of the fungal genomic DNA extraction kit, and use 16 Cordyceps sinensis culture fluid genomic DNA as templates to carry out nested PCR amplification (same as step 3 in Example 1) to detect the Cordyceps sinensis culture fluid Presence of Paecilomyces chrysalis in . The detection result shows that in 16 Cordyceps sinensis culture fluids, Paecilomyces paecilomyces was detected in No. 5 and No. 13 bottles, and the detection primer set and detection method provided by the present invention can well detect Paecilo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com