Specific PCR (polymerase chain reaction) method for rapidly identifying common high temperature actinomyces
A high-temperature actinomycetes, common technology, applied in the field of molecular biology identification, can solve the problems of cumbersome and time-consuming identification of conventional technology, unable to meet the requirements of common high-temperature actinomycetes screening and application research, etc., to achieve convenient cost, primer specificity Strong performance and simple PCR amplification conditions
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0026] Example 1: Design of specific PCR primers
[0027] According to the common high temperature actinomycetes registered on GenBank gyrB Gene sequence information for primers
[0028] design (see attached figure 1 ).
[0029] The GenBank accession number of the reference sequence is: AB242203 ( Thermoactinomyces vulgaris , gyrB gene, complete cds, http: / / www.ncbi.nlm.nih.gov / nuccore / AB242203.1).
[0030] An upstream primer was designed at 89-109 and named 109F. The sequence is: 5'-GCGTGACGGGAAAATCTATC-3'.
[0031] A downstream primer was designed at 800-822 and named 801R. The sequence is: 5'-CATCACGGCTTTGTTAATAATC-3'.
Embodiment 2
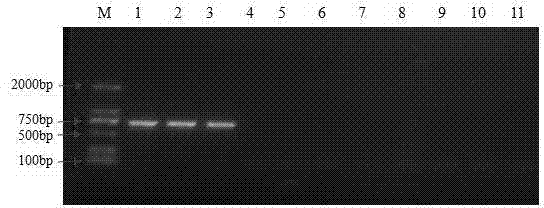
[0032] Example 2: Primer Specificity Verification
[0033] 1.1 Strains of actinomycetes common and their relatives
[0034] common high temperature actinomycetes ( Thermoactinomyces vulgaris DSM 43016), Thermoactinomycetes intermediate ( Thermoactinomyces intermedius DSM 43816), Laysia saccharum ( Laceyella sacchari DSM 43356), Laceyella putida ( Laceyella putidus DSM44608) was purchased from the German Culture Collection of Microorganisms; Bacillus thuringiensis ( Bacillus thuringiensis CICC 22945), Bacillus licheniformis ( Bacillus licheniformis CICC 10101) from China Industrial Microorganism Culture Collection Management Center; Laceyella tengchongensis CCTCC AA 208050, Laceyella sediminis CCTCC AA 2011024 was purchased from China Type Culture Collection Center; Thermoactinomyces vulgaris ACCC 41061) was purchased from the China Agricultural Microorganism Culture Collection Management Center; Thermoactinomyces vulgaris ZM60) was isolated and identifi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com