Specific Molecular Detection Primers for Soybean Gray Spot and Its Application
A technology for gray spot disease and soybean, which is applied in the field of specific molecular detection primers for soybean gray leaf spot bacteria, can solve the problems of high similarity, inability to design specific detection primers, etc., and achieves strong specificity, simple and fast method, and high sensitivity. Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0021] Establishment of a specific molecular detection method for Soybean gray spot fungus
[0022] 1. Primer Design
[0023] The most commonly used ITS sequences in the molecular detection of plant pathogenic fungi have high similarities among some fungal species, and it is difficult to directly use them for accurate species identification. In the present invention, through the multiple alignment analysis of ITS, β-tublin, Actin, Cytb, EF-1α and other gene sequences of Soybean gray spot and other plant pathogenic fungi, multiple soybean gray spots are finally found in the β-tublin gene sequence Pathogen-specific loci, and based on this, specific detection primers for Soybean gray spot were designed and screened. The β-tublin gene sequences of Cercospora sojina and other fungi of the genus Cercospora were downloaded from the GenBank database, and multiple alignment analysis was performed using MEGA 5.1 software, and molecular detection primers were designed for the specific s...
Embodiment 2
[0034] Specific Validation of Primers for Detection of Gray Spot of Soybean
[0035] 1. Extraction of genomic DNA from the sample to be tested
[0036] Genomic DNA of soybean sclerotinia, soybean purple spot, soybean Phytophthora root rot, soybean scab and soybean blight were extracted by the above-mentioned SDS method.
[0037] 2. Specific detection of primers for detection of gray spot disease of soybean
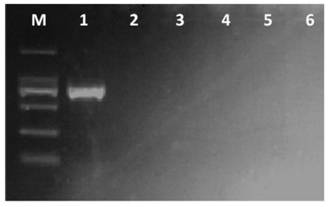
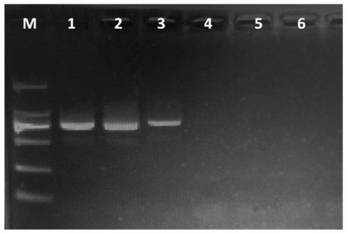
[0038]PCR amplification was carried out using the genomic DNA of the above-mentioned tested fungi as templates. The PCR reaction system contains: 2.5 μL 10×PCR buffer, 1.5 μL Mg 2+ (25mM), 0.5μL dNTP (each 10mM), 2.5U Taq DNA polymerase, 1.0μL each of upstream primer TF1 (10μM) and downstream primer TR1 (10μM), 1.5μL DNA template, ddH 2 O 16.5 μL. The PCR reaction program was: 94°C for 3min; 35 cycles of 94°C for 30s, 57°C for 30s, 72°C for 1min; 72°C for 10min. 5.0 μL of the PCR product was electrophoresed on a 1.5% (m / v) agarose gel, stained with ethidium bromide, a...
Embodiment 3
[0042] Sensitivity detection of detection primers for soybean gray spot bacterium
[0043] 1. Extraction of Genomic DNA of Soybean Gray Spot
[0044] Genomic DNA of Soybean gray spot was extracted by the above-mentioned SDS method
[0045] 2. DNA concentration determination and dilution
[0046] Use a NanoDrop ultra-micro spectrophotometer to measure the concentration of the above-mentioned genomic DNA, and use ddH 2 Oy was serially diluted to 100ng / μL, 10ng / μL, 1ng / μL, 10 -1 ng / μL, 10 -2 ng / μL and 10 -3 ng / μL.
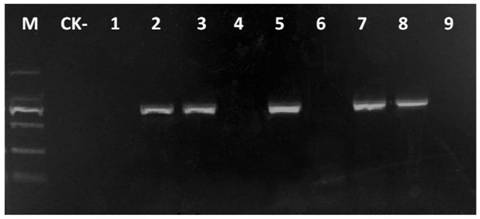
[0047] 3. Perform PCR amplification using the above-mentioned different concentrations of DNA as templates. The PCR reaction system contains: 2.5 μL 10×PCR buffer, 1.5 μL Mg 2+ (25mM), 0.5μL dNTP (each 10mM), 2.5U Taq DNA polymerase, 1.0μL each of upstream primer TF1 (10μM) and downstream primer TR1 (10μM), 1.5μL DNA template, ddH 2 O 16.5 μL. The PCR reaction program was: 94°C for 3min; 35 cycles of 94°C for 30s, 57°C for 30s, 72°C for 1min; 72°C for 10min. ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com