Molecular marker for identifying trichotylenchus changlingensis and application thereof
A molecular marker, nematode technology, applied in the determination/inspection of microorganisms, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problems of high technical requirements and poor identification effect, achieve high identification accuracy, simple detection method, Primer-specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
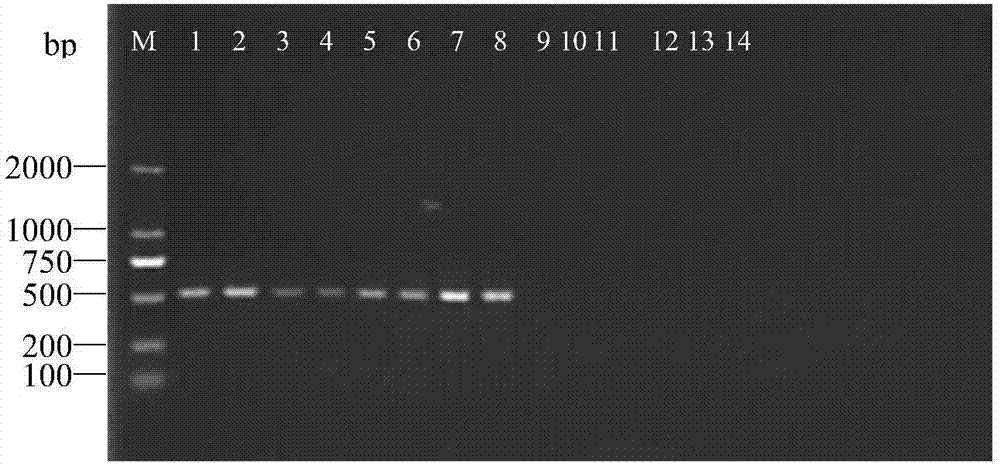
[0045] Example 1. Design of specific primers for C. changling according to the present invention and PCR detection test of primer specificity
[0046] Proceed as follows:
[0047] (1) Test materials and sources
[0048] 1. C. elegans hairpad: 8 HEB1, HEB2, HEB3, SY1, CC1, QD1, ZB1 and XT1 in total.
[0049] 2. Other nematodes: P. elegans C1, D. nematode D1, Root-knot nematode M1, Cyst nematode H1 and Brachypodium nematode P1.
[0050] The above nematodes were preserved in the Institute of Plant Protection, Hebei Academy of Agriculture and Forestry.
[0051] (2) Test method
[0052] (1) Single nematode DNA extraction. The nematodes were washed in sterile deionized water, and a single nematode was picked and placed in 10 μL of lysis buffer (1×PCR Buffer (Mg 2+ ), 1 μg proteinase K), water bath at 65 °C for 1 min, liquid nitrogen treatment for 2 min, after repeated freezing and thawing several times, incubate at 56 °C for 20 min, and heat at 95 °C for 10 min to obtain DNA ex...
Embodiment 2
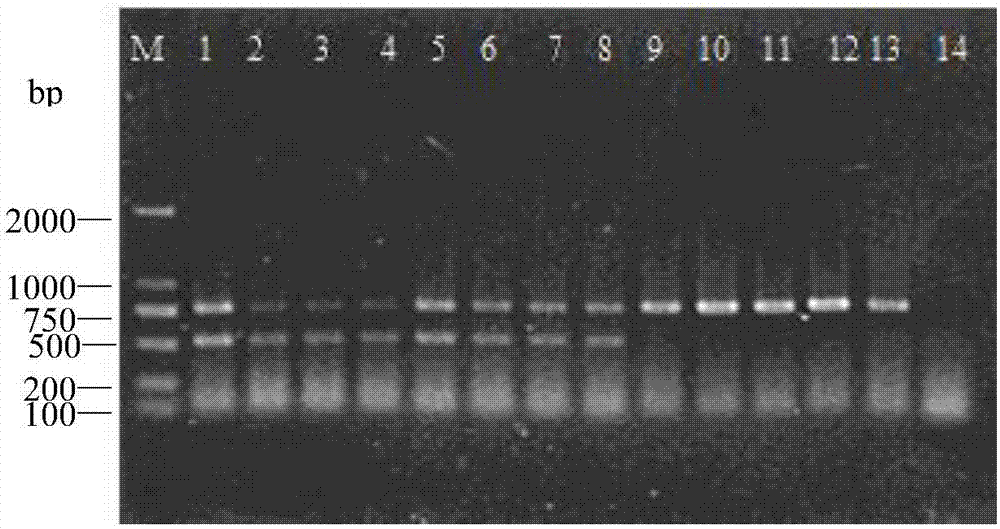
[0062] Embodiment 2, the double PCR method of the present invention detects the specificity test of C. elegans
[0063] (1) Test material: the same as (1) of Example 1.
[0064] (2) Test method:
[0065] (1). Single nematode DNA extraction. Same as (2) (1) of Example 1.
[0066] (2). Double PCR amplification. Using nematode DNA as a template and TC-F1 / TC-R1 and D2A / D3B as primers, double PCR amplification was performed to obtain the amplified product. The PCR reaction system was 25 μL: 10×Ex Taq Buffer (Mg 2+ ) 2.5μL, 5U / μL Ex Taq polymerase 0.2μL, template DNA 0.5μL, 2.5mmol / L dNTPs Mixture 2μL, primers TC-F1 / TC-R1 0.5μL each, D2A / D3B (10μmol·L -1 ) each 1 μL, and finally make up ddH2O to 25 μL, and set a negative control without template. PCR reaction program: 95°C for 4 min; 94°C for 30s, 62°C for 45s, 72°C for 45s, 35 cycles; 72°C for 10 min; storage at 16°C. The primers D2A and D3B described therein are universal primers in the 28S region, which are used as interna...
Embodiment 3
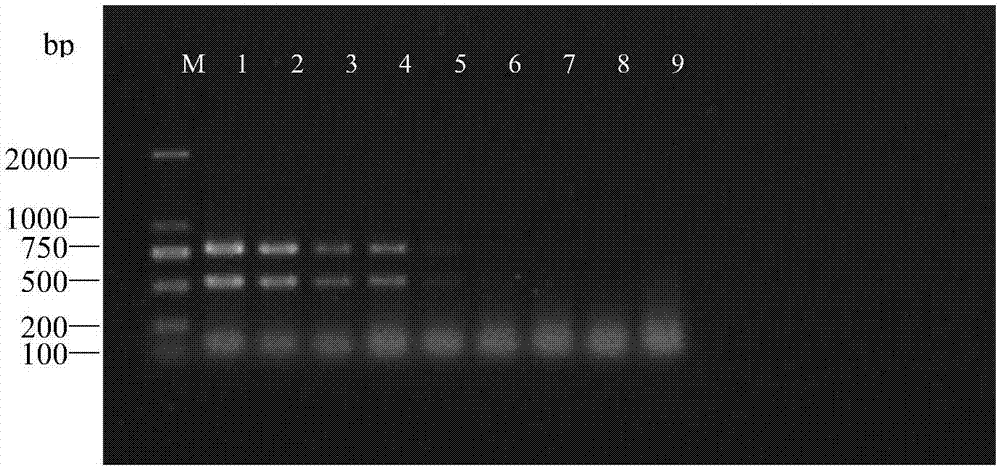
[0072] Embodiment 3, the detection sensitivity test of the double PCR method of the present invention
[0073] (1) Test materials
[0074] The single nematode DNA was diluted in a concentration gradient of 1 / 4×, 1 / 8×, 1 / 16×, 1 / 32×, 1 / 64×, 1 / 128×, 1 / 256×, and 1 / 512×.
[0075] (2) Test method
[0076] (1). The extraction method of HEB1 single nematode DNA is the same as that in Example 1.
[0077] (2). The double PCR amplification method is the same as that in Example 2.
[0078] (3). Electrophoresis detection is the same as in Example 1.
[0079] result (see image 3 ) single nematode DNA is 1 / 4× (concentration 1 ng / μL) to 1 / 32× (concentration 0.125ng / μL) (see image 3 Swimming lanes 1-4), clear and stable bands can be amplified, that is, detection can be performed at a very low template concentration of 0.125ng / μL, indicating that the double PCR detection method of the present invention has high sensitivity.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com