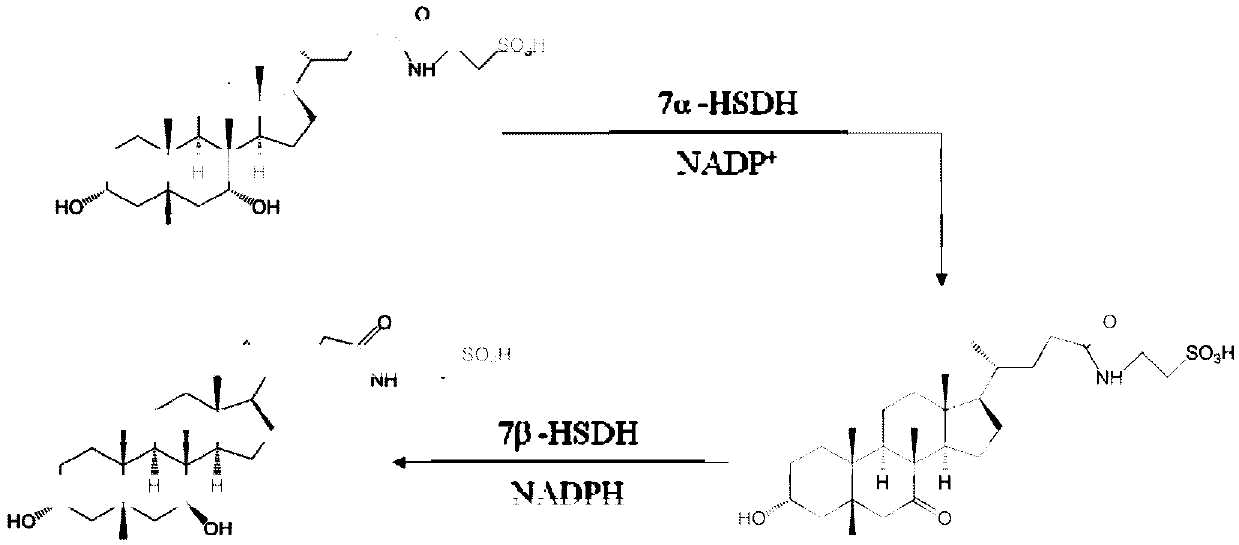
7 alpha-hydroxysteroid dehydrogenase as well as coding gene and application thereof
A technology for encoding genes and residues, applied in the fields of application, genetic engineering, plant genetic improvement, etc., can solve the problem that the activity and stability of hydroxysteroid dehydrogenase cannot meet the needs of industrial production at the same time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
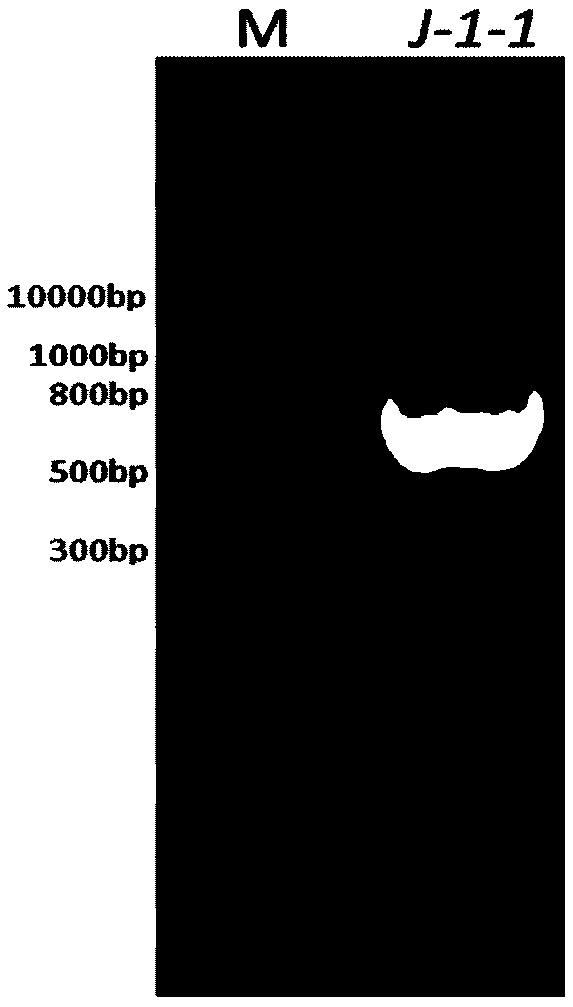
[0057] Example 1, Discovery of a new 7α-HSDHJ-1-1 gene
[0058] Fecal samples from healthy black bears in the Sichuan Black Bear Conservation and Incubation Base were collected using sterilized medicine spoons, stored on dry ice and transported back to the laboratory. The total DNA of black bear feces was extracted using the Qiagen Fecal DNA Genome Extraction Kit, and submitted to Shanghai Meiji Biomedical Technology Co., Ltd. for metagenomic sequencing. Comparison of the existing Clostridium sardinica 7α-hydroxysteroid dehydrogenase (Clostridium sardinia 7α-HSDH) gene sequence with metagenomic sequencing data revealed a novel 7α-HSDHJ-1-1 Gene, the nucleotide sequence of which is shown in SEQ ID NO.2, wherein the sequence 2 in the sequence table is an open reading frame from the 1st position to the 786th position at the 5' end, and the open reading frame part is 786bp, and the 7α - The amino acids of the HSDHJ-1-1 protein are shown in SEQ ID No. 1, and the sequence 1 consist...
Embodiment 2
[0059] Embodiment 2, the cloning of gene
[0060] 1) Primer design: design primers for the new 7α-HSDHJ-1-1 gene sequence obtained by sequencing, and the nucleotide sequences of the primers are as follows:
[0061] J-1-1-BamHI-F: 5'-CGGGATCCATGAGAGTAAAAGATAAAATAG-3' (sequence 3 in the sequence listing),
[0062] J-1-1-XhoI-R: 5'-CGCTCGAGTTATCTTTGAACAAGGTCTCCGTATT-3' (SEQ ID NO: 4 in the Sequence Listing).
[0063] 2) PCR amplification: the total DNA of black bear feces was used as a template, and the primers obtained in step 1) were used to amplify according to the following PCR system and procedure.
[0064] PCR system includes: template DNA is 0.5 μL, J-1-1-BamHI-F (10 μM) is 2 μL, J-1-1-XhoI-R (10 μM) is 2 μL, 0.1% BSA is 3 μL, PrimeSTAR HS DNA Polymerase It is 25μL, add ddH 2 O to PCR system is 50 μL.
[0065] The PCR program is as follows:
[0066] a. Pre-denaturation at 94°C for 5 minutes;
[0067] b. Denaturation at 98°C for 10 sec, annealing at 53°C for 10 sec, e...
Embodiment 3
[0076] Embodiment 3, expression, extraction and purification of new gene
[0077] 1. Construction of the connection vector: the full-length sequence of the 7α-HSDHJ-1-1 gene was connected to the vector pGEX-6p-2.
[0078] The ligation system includes: 6 μL for the double-digested 7α-HSDHJ-1-1 gene, 1 μL for T4DNALigase, 2 μL for the vector pGEX-6p-2, 1 μL for 10×T4Ligase buffer, 10 μL for the system, and 16 °C for 12 hours.
[0079] 2. To transform E.coli DH5α competent cells, the steps are as follows:
[0080] 1) Competent cells E.coli DH5α (purchased from Tiangen Biochemical Technology (Beijing) Co., Ltd., item number: CB101) were placed on ice to thaw.
[0081] 2) Add the connection system obtained in step 1 into the melted E.coli DH5α competent, and keep it on ice for 30 minutes.
[0082] 3) Heat treatment at 42°C for 90s.
[0083] 4) Stand on ice for 2 minutes.
[0084] 5) Add 600 μL of anti-antibody-free LB medium, shaker temperature 37° C., shaker speed 150 rpm, tim...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com