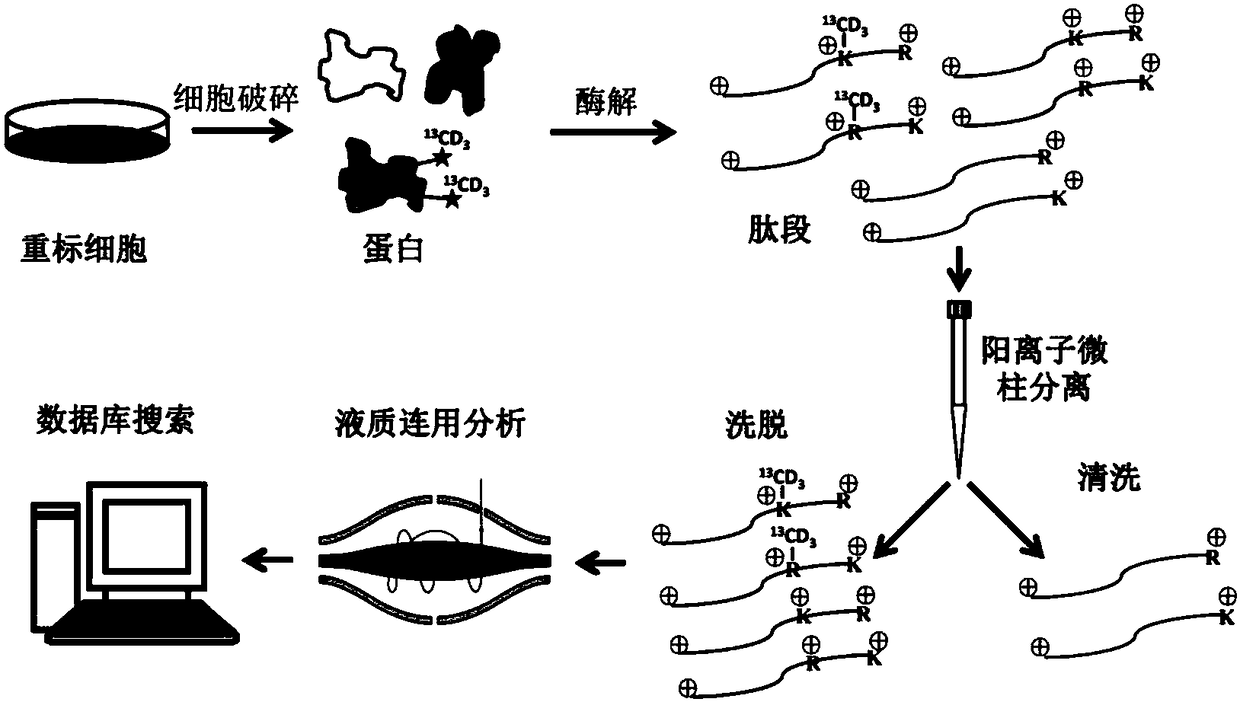
Analytic treatment method for protein methylation and application thereof
A technology for analytical processing and methylation, applied in the field of analysis and processing of protein methylation, can solve problems affecting the specificity of separation, efficient separation of methylated peptides and interference
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
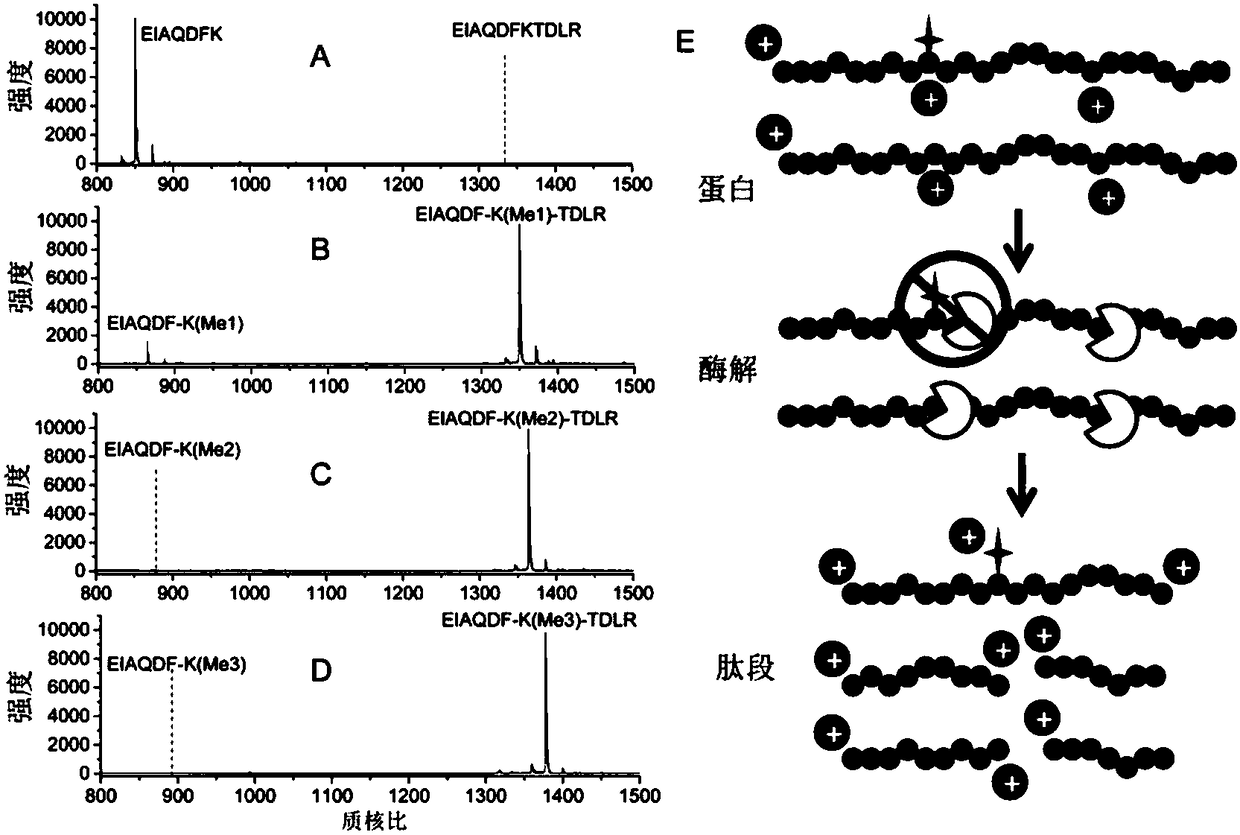
[0032] Investigation of the missing cutting situation of methylation sites: unmodified (EIAQDF-K-TDLR), monomethylated (EIAQDF-K(Me1)-TDLR), dimethylated (EIAQDF-K(Me2)-TDLR ) and trimethylated (EIAQDF-K(Me3)-TDLR) peptides dissolved in 50mM NH 4 HCO 3 (pH 8.0), add trypsin according to the ratio of protein: enzyme (50:1, w / w), incubate at 37°C for 16 hours, and then perform laser-assisted ionization time-of-flight mass spectrometry (MALDI-TOF) analysis. Such as figure 2 As shown, by analyzing the mass spectrograms of different peptides after trypsin digestion, it can be found that the C-terminus of unmodified lysine will be completely cut off, and the C-terminus of monomethylated lysine Only a small proportion of the carbon terminal (C-terminal) will be digested, and the C-terminal of lysine modified by dimethylation and trimethylation cannot be digested by trypsin. This result clearly shows that the carbon terminus (C terminus) of the methylated lysine residue and argini...
Embodiment 2
[0034] The multi-enzyme digestion strategy is used to reduce the proportion of missed cleavage sites: add 10-20mM dithiothreitol (DTT) at a final concentration of 1mg protein sample extracted from HepG2 cells, 1- 3h, then add iodoacetamide (IAA) at a final concentration of 20-40mM, and react in the dark at 20-25°C for 40-60min. The solution was replaced with 50 mM ammonium bicarbonate (pH 8.0) by an ultrafiltration assisted sample processing strategy. Divide 1 mg of the treated protein sample into three parts. The first group of samples was added with trypsin according to the ratio of protein: enzyme (50:1, w / w), and incubated at 37°C for 16 hours; the second group of samples was Add trypsin (trypsin) and lysinase (lys-C) at the ratio of protein: enzyme (50:1, w / w), and incubate at 37°C for 16 hours; / w) was added to trypsin and lysinase (lys-C), and incubated at 37°C for 16 hours. Remove trypsin (trypsin) and lysinase (lys-C) by ultrafiltration, then add 10× activation solu...
Embodiment 3
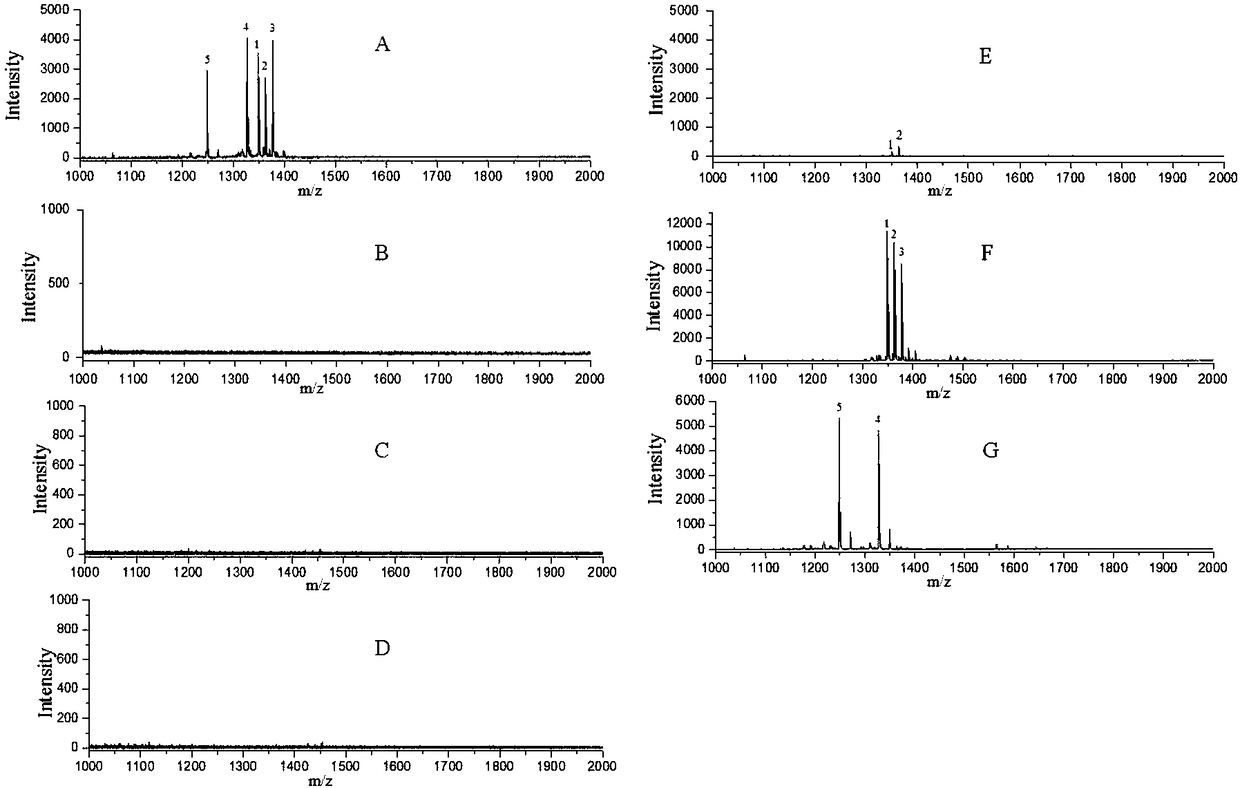
[0041] Optimization of separation conditions for "SCXtip": Use methylated standard peptides to investigate the optimal separation conditions required for SCXtip separation. 5 methylated standard peptides (EIAQDF-K(Me1)-TDLR; EIAQDF-K(Me2)-TDLR; EIAQDF-K(Me3)-TDLR; SG-R(Me1)-GGNFGFGDSR; N-R(Me2S)-GAGGFGGGGGTR ) was dissolved in 60% (vol / vol) acetonitrile, 40% (vol / vol) 5mM BRUB (pH 2.5) solution, loaded into the SCXtip, and successively with 90% acetonitrile, 5mM BRUB (pH9) solution, 85% Acetonitrile, 5mM BRUB (pH 9) solution, 80% acetonitrile, 5mM BRUB (pH 9) solution, 65% acetonitrile, 5mM BRUB (pH 9) solution, 30% acetonitrile, 5mM BRUB (pH 12) solution bound to SCXtip column Peptides were eluted, and the corresponding elution solutions were collected and analyzed by MALDI-TOF mass spectrometry. image 3 , we can see that in the mass spectrum of the sample loading step, no peptide peaks are seen, indicating that the methylated peptides are all retained on the SCXtip; when d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com