Method for detecting bacterial fruit blotch and xanthomonas malvacearum of melons simultaneously and special kit thereof
A kind of technology of fruit spot bacteria and angular spot bacteria, which is applied in the biological field
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
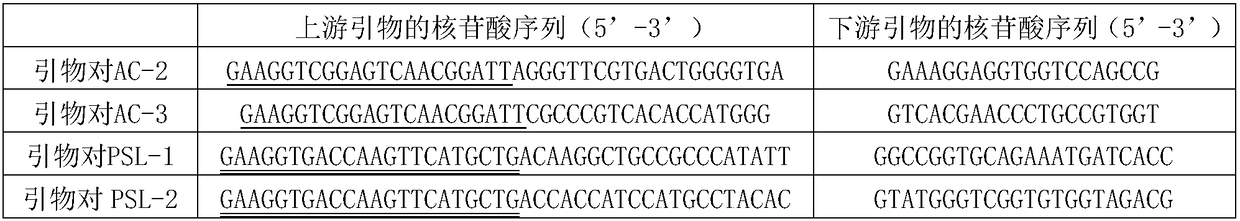
[0103] Embodiment 1, the synthesis of primer
[0104] According to the nucleotide sequence of the par gene in the AC bacteria (shown as sequence 1 in the sequence listing), primers were artificially designed and primer pair AC-2 and primer pair AC-3 were synthesized. According to the nucleotide sequence of the gap1 gene in the PSL bacterium (shown in sequence 2 in the sequence table), artificially design and synthesize primers to PSL-1, primers to PSL-2, primers to PSL-3, primers to PSL-4, primers Pair PSL-5, primer pair PSL-6 and primer pair PSL-7. Primer design principles: the length of each primer is 20-26bp, and the size of the amplified fragment is about 100bp. TM The Tm value is adjusted in the software to be 55-62°C; the upstream primers of the primer pair (i.e., primer pair AC-2 and primer pair AC-3) designed according to the AC bacteria all contain linker sequence 1 (linker sequence 1 can bind to the FAM signal gene group, thus being Kraken TM software identificati...
Embodiment 2
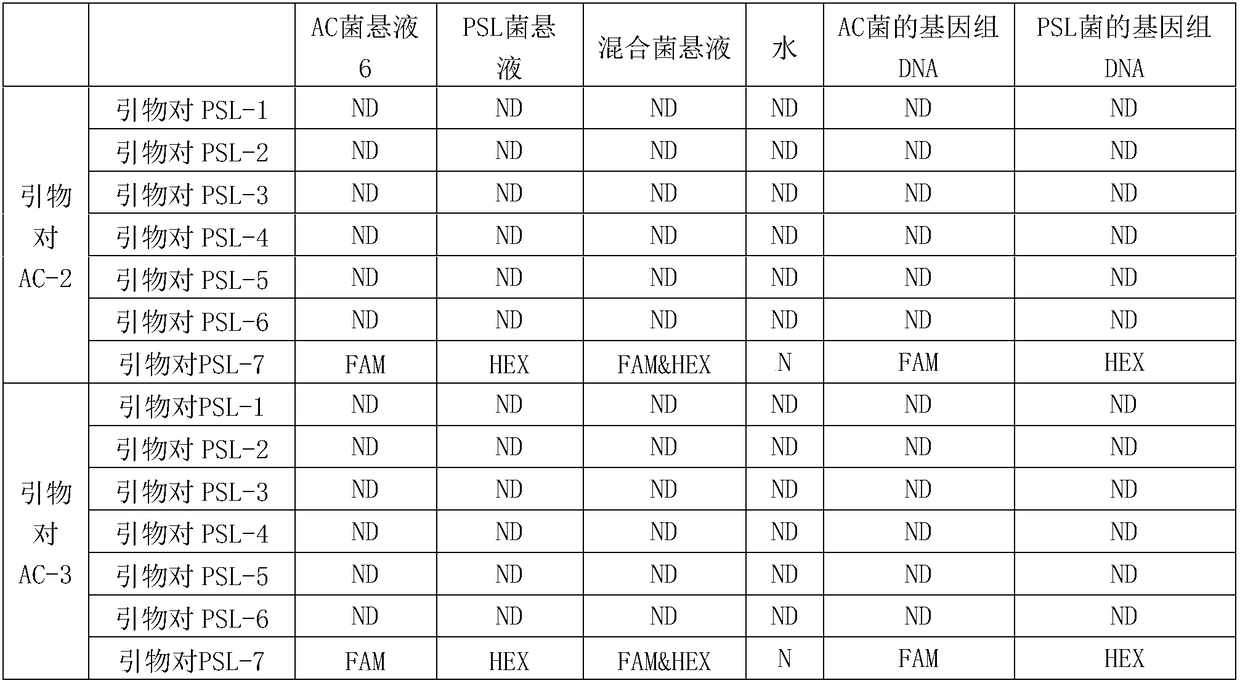
[0110] Embodiment 2, the screening of the primer group that detects AC bacterium and PSL bacterium
[0111] 1. Screening of primers for the detection of AC bacteria
[0112] 1. Obtaining the template
[0113] Take the bacterial suspension of AC bacteria and add sterile water to obtain a concentration of 10 8 CFU / mL of AC bacterial suspension 6 (OD 600nm is 0.15).
[0114] Take the AC bacterial suspension 6, and carry out gradient dilution with sterile water to obtain a concentration of 10 3 CFU / mL AC bacterial suspension 1, the concentration is 10 4 CFU / mL AC bacterial suspension 2, the concentration is 10 5 CFU / mL AC bacterial suspension 3, the concentration is 10 6 CFU / mL of AC bacterial suspension 4 and concentration of 10 7 CFU / mL of AC bacterial suspension 5.
[0115] 2. Obtaining the reaction mixture
[0116] 12 μL of the upstream primer of primer pair AC-2 (at a concentration of 10 mM) and 15 μL of the downstream primer of primer pair AC-2 (at a concentration o...
Embodiment 3
[0148] The test of the primer group 1 and the primer group 2 of embodiment 3, embodiment 2 screening
[0149] 1. AC bacteria and PSL bacteria are mixed in different proportions
[0150] 1. Obtaining the template
[0151] Mix the AC bacterial suspension 6 and the PSL bacterial suspension (see the first column of Table 4 for the volume ratio) to obtain the mixed bacterial suspension, that is, the template.
[0152] 2. Obtaining the reaction mixture
[0153] Mix 12 µL of the upstream primer of primer pair AC-2 (at a concentration of 10 mM), 15 µL of the downstream primer of primer pair AC-2 (at a concentration of 10 mM), 12 µL of the upstream primer of primer pair PSL-7 (at a concentration of 10 mM), and 15 µL of the upstream primer of primer pair PSL-7 -7 downstream primers (10 mM concentration) were mixed to obtain primer mixture a.
[0154] Mix 12 µL of the upstream primer of primer pair AC-3 (at a concentration of 10 mM), 15 µL of the downstream primer of primer pair AC-3 (a...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com