Pathogen biomarkers and uses therefor
A technology of biomarkers and indicators, applied in the field of diagnosis and/or monitoring of virus-related systemic inflammation, peripheral blood RNA and protein biomarkers, which can solve the problems of making decisions about the treatment and management of clinically diagnosed patients
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
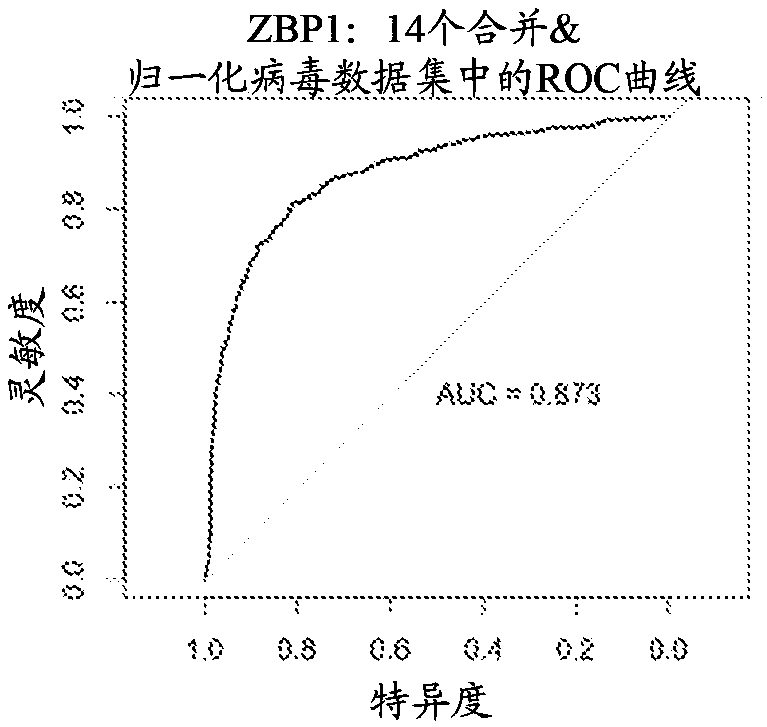
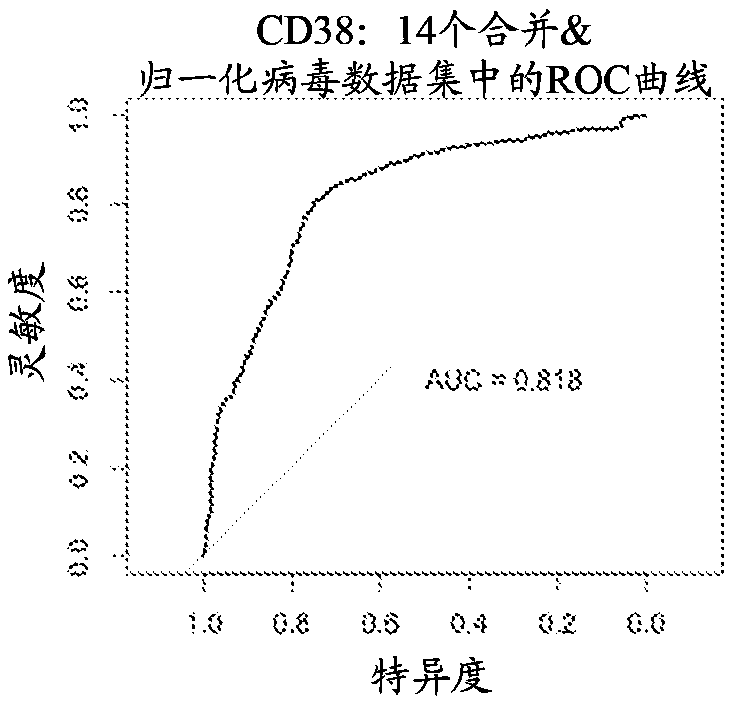
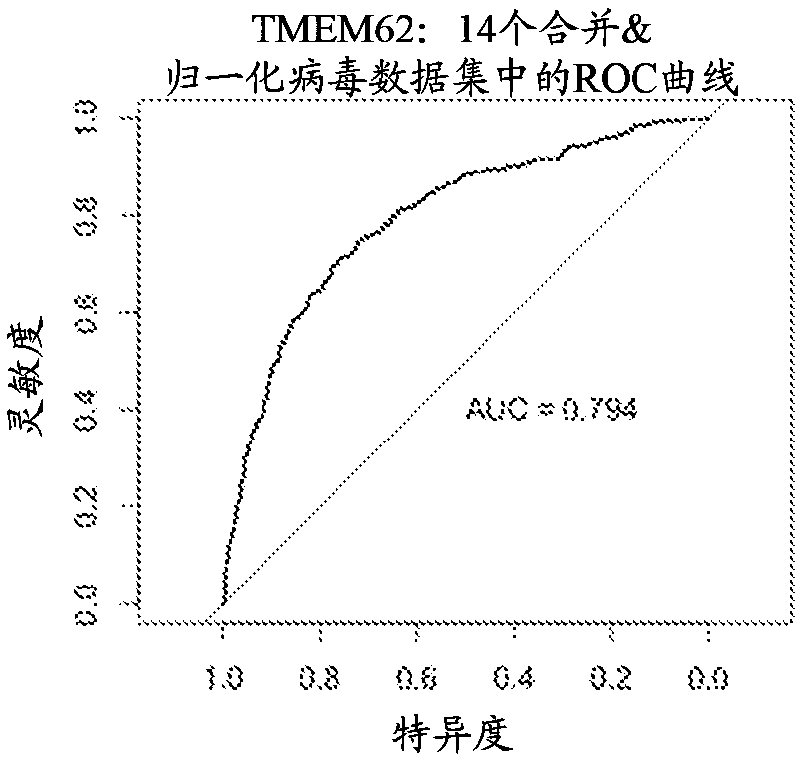
[0198] Group A biomarkers and their diagnostic performance across multiple data sets
[0199] Three separate biomarkers were found to have strong diagnostic performance across all virus data sets (as measured using area under the curve (AUC)). The three separate biomarkers include ZBP1 (DNA SEQ ID NO: 399) , TMEM62 (DNA SEQ ID NO: 414) and CD38 (DNA SEQ ID NO: 415). Four "core virus" data sets (including dengue fever, Lassa fever, influenza and herpes virus infection) were used to discover these biomarkers. Then ten "validating viruses" data sets are used to validate these biomarkers. These data sets include samples collected from patients suffering from a wide range of viral infections, including adenovirus, herpes virus, enterovirus, rhinovirus, hepatitis C virus, measles, rotavirus, and respiratory syncytial virus. The "non-viral inflammation" dataset includes conditions that cause systemic inflammation, including, among others, bacterial infections, allergic reactions, and ...
Embodiment 2
[0201] Groups B, C, D and E biomarkers. Group the biomarkers based on their correlation with ISG15, IL16, OASL and CD97
[0202] Two pairs of derived biomarkers (IL16 / ISG15; CD97 / OASL) were found to provide the highest AUC across all virus data sets studied. Then AUC will be provided> 0.8 (see Table 13 for the complete list of derived biomarkers) as ratio biomarkers based on their correlation with ISG15 (group B), IL16 (group C), OASL (group D) or CD97 (group E) Sex (as presented in Table 10) was assigned as a separate biomarker to one of the four groups.
Embodiment 3
[0204] VASIRS biomarker export (individual and exported biomarkers)
[0205] An illustrative process for identifying VaSIRS biomarkers for use in diagnostic algorithms will now be described.
[0206] Gene expression data (from clinical trials conducted by the inventors or from Gene Expression Omnibus) were analyzed using various statistical methods to identify individual and derived biomarkers (ratio), but mainly followed the method described in WO2015 / 117204. The individual and derived markers are ranked based on performance (area under the curve).
[0207] The data set used for training is derived from GEO (all comply with MIAME) and has the following limitations: use peripheral blood samples, use appropriate controls, use appropriate number of samples to provide significance after adjusting for false discovery rate (FDR), all The data passed standard quality control measures, and the principal component analysis did not show any artifacts or potential bias. The data set is divid...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com