Rapid detection kit for identifying SNP characteristic sites of Y chromosome haplotype pedigree and method
A technology of Y chromosome and characteristic bits, applied in the field of detection, can solve problems such as difficult large-scale promotion and use, complex library construction and sequencing process, difficult high-throughput rapid detection, etc., to save experimental costs, simplify experimental procedures, Workload saving effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0092] D-JST021355 (rs2267802)
[0093] rs2267802-F: 5'-CCCAGCCCATTTTATCTATCT-3'
[0094] rs2267802-R: 5'-TCAGGGTAGAAATTAAAAGAGGA-3'
[0095] Probe is forward:
[0096]rs2267802-P-A AGAAACCTCTAAACCTA 5`6-FAM, 3`BHQ-X
[0097] rs2267802-P-G AGAAACCTCTGAACCT 5`HEX, 3`BHQ-X
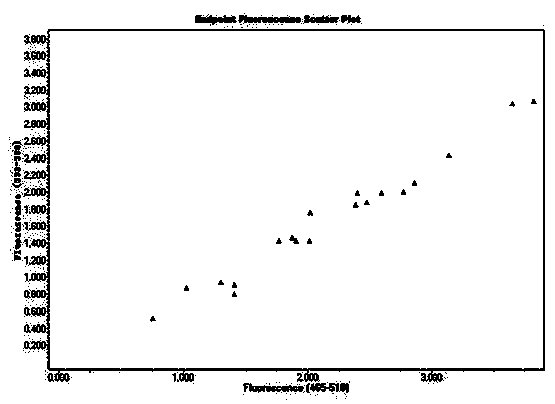
[0098] Type A is the MP type probe, labeled with FAM; blue fluorescence, manifested as type X
[0099] G-type is the HP-type probe, which is HEX-labeled; green fluorescence, which is Y-type
Embodiment 2
[0101] D1-N1 (rs3212291)
[0102] rs3212291-F: 5'-ATCTGCTAGCAAAGTTGGCTT-3'
[0103] rs3212291-R:5'-CTATTCAAGCTACTACCTTAAAGCA-3'
[0104] Probe for reverse:
[0105] rs3212291-P-C CCAGGAGAAAATC 5`6-FAM, 3`BHQ-X
[0106] rs3212291-P-G CCAGGACAAAATC 5`HEX, 3`BHQ-X
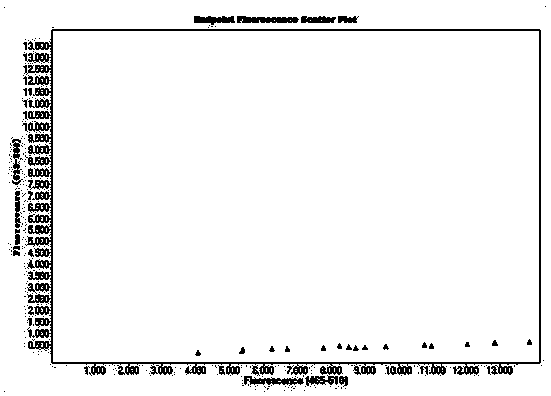
[0107] C-type is MP-type probe, labeled with FAM; blue fluorescence, manifested as X-type
[0108] G-type is the HP-type probe, which is HEX-labeled; green fluorescence, which is Y-type
Embodiment 3
[0110] E-M96(rs9306841)
[0111] rs9306841-F: 5'-CCATATATTTTGCCATAGGTTTT-3'
[0112] rs9306841-R:5'-CTGTGATGTGTAACTTGGAAAAC-3'
[0113] Probe for reverse:
[0114] rs9306841-P-C CTCATAATAGGATAAAAC 5`6-FAM, 3`BHQ-X
[0115] rs9306841-P-G CTCATAATACGATAAAAC 5`HEX, 3`BHQ-X
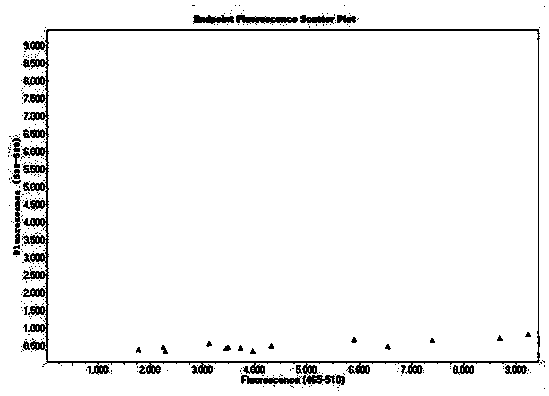
[0116] C-type is MP-type probe, labeled with FAM; blue fluorescence, manifested as X-type
[0117] G-type is the HP-type probe, which is HEX-labeled; green fluorescence, which is Y-type
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com