Quantitative measurement of hepatitis b virus cccdna
A technology of DNA molecules and DNA sequences, applied in the field of quantitative measurement of hepatitis B virus CCCDNA, can solve the problems of high cost and side effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0095] Example 1: Recovery of DNA after bisulfite treatment as detected by PCR.
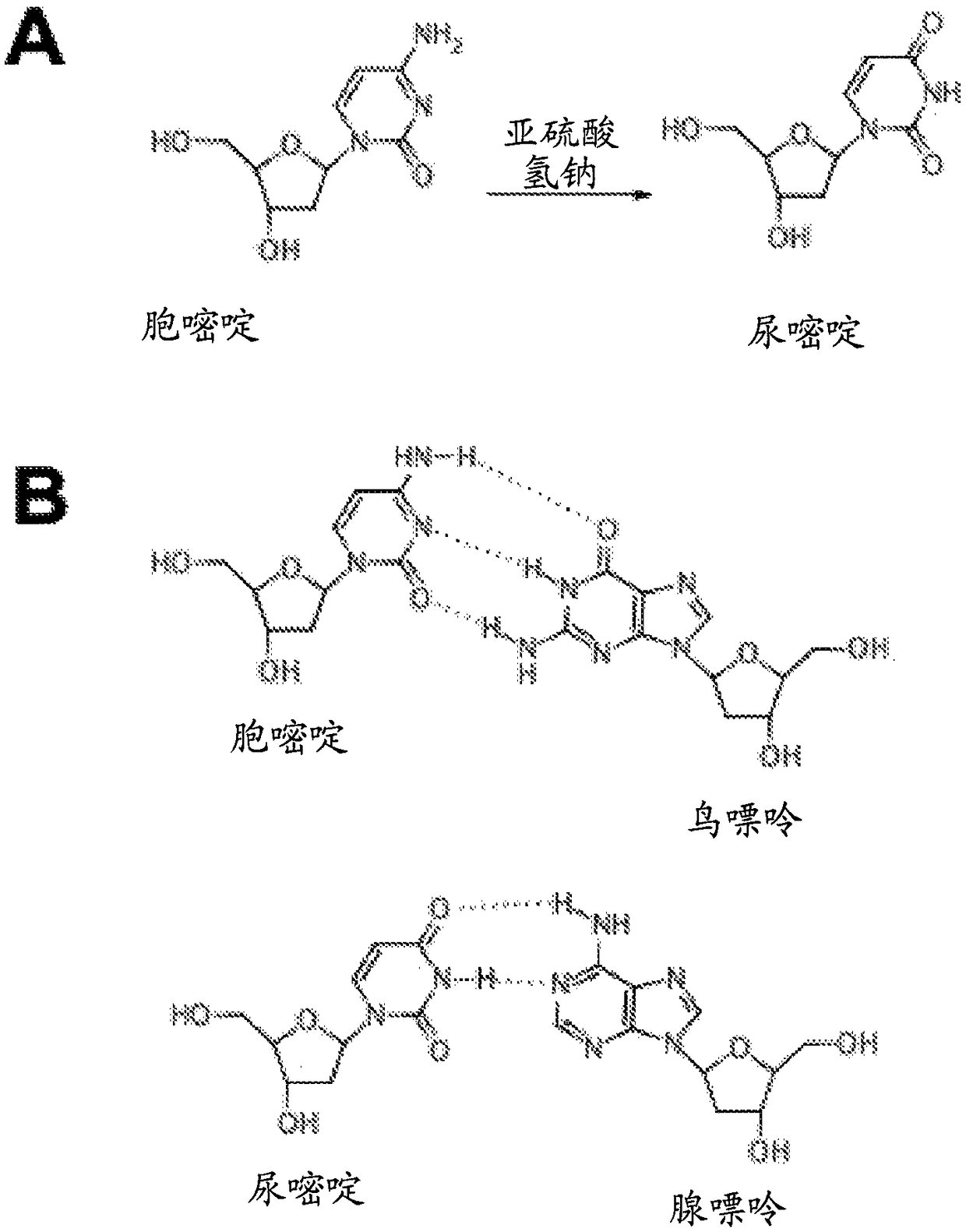
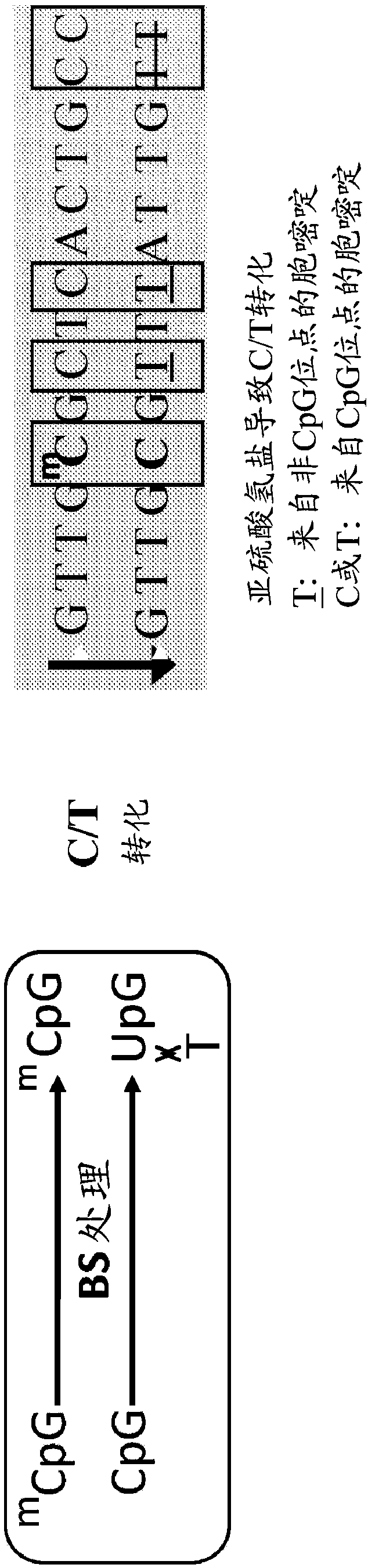
[0096] The complementary strands of double-stranded DNA are rendered non-complementary by bisulfite treatment, which converts cytosine nucleotides to uracil, as Figure 2B shown.
[0097] Here, the Qiagen Epitect Bisulfite Conversion Kit (Qiagen, CA) and the Zymo Research EZ DNA Methylation Gold Kit (ZymoResearch, Zymo Research Corporation, CA) were used according to the manufacturer's instructions for bisulfite conversion. DNA from human HCC tissue samples was bisulfite-treated and tested with specific assays before and after bisulfite treatment. Regions on the constitutive gene actin were used for comparison by running assays targeting non-bisulfite-treated DNA regions and bisulfite-treated DNA regions (see Figure 8 ).
[0098] Figure 8 It was shown that bisulfite treatment (BS) did not significantly reduce copy detection of DNA target regions. Primers for the amplification of short prod...
Embodiment 2
[0100] Example 2: DNA quantification by real-time polymerase chain reaction (RT-PCR).
[0101] Total HBV DNA was quantified by real-time PCR using the LightCycler PCR instrument (Roche Biochemical, Germany) and LightCycler Probe Master Mix (Roche Biochemical, Germany) according to the manufacturer's instructions. Primers for bisulfite-treated total HBV DNA (forward, 5'-TTATGTTAATGTTAATATGGGTTTAAA-3'; reverse, 5'-TTCTCTTCCAAAAATAAAACAA-3'; probe, 5'-6FAM-TTAGATAATTATTG+TGG+T+ T+T+TA+TA+T-BBQ 3') and serially diluted HBV DNA plasmids were used as quantitative standards for quantification.
[0102] To quantify bisulfite-converted cccDNA, primers within the gap region of the (+) strand (forward, 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTTTGTYGGATYGTGTGTATTT-3′; reverse, 5′-AACRTTCACRATAATCTCCATAC-3′) were selected to prevent rcDNA amplification. TaqMan probe 5'6FAM-TTTTAT+T+T+T+T+G+TAY+G+TA-BBQ-3' was used to detect the amplification product.
Embodiment 3
[0103] Example 3: 1-step BS-cccDNA assay for detection of cccDNA by real-time PCR.
[0104] Figure 7 shows the sensitivity of 100 copies of BS-cccDNA template in a one-step cccDNA assay, and the sensitivity at 10 5 10 copies of rcDNA background 8 Linearity of synthetic BS-cccDNA to 100 copies. Figure 5 The 10-fold serial dilutions of the synthetic HBV template described in range from 100 to 10 for the sensitivity and linearity of the assay. 8 copies. Data are presented as amplification curve (top) and standard curve (bottom).
[0105] One-step real-time PCR was performed using the primer set of SEQ ID NO: 11 and SEQ ID NO: 24. Detection of the amplified product was performed using SEQ ID NO: 29 as a probe. Human methylated bisulfite-treated DNA was used as a negative control at 10,000 copies and showed no observable amplification. Note, 10 4 HMBS are amplified at cp values similar to 10 copies of cccDNA, so at 10 4 The sensitivity or limit for detection of 1-step ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com