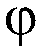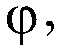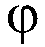Methods for obtaining and correcting biological sequence information
A technology of sequence information and sequencing, which is applied in the fields of bioinformatics, biochemical equipment and methods, and the determination/inspection of microorganisms, and can solve problems such as no sequencing logic innovation.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0376] Example 1: Sequencing by "2+2 monochrome" method
[0377] In order to further describe the present disclosure, specific examples are provided below. Unless otherwise indicated, specific parameters, procedures, etc. are conventional in the art. The specific examples are not intended to limit the scope of the invention.
[0378] For sequencing by the "2+2 monochrome" method, three sets of reactions were prepared. Each set includes two vials, each containing two bases labeled with the same fluorophore X. For each set, two vials contained exactly all four bases of the sequencing reaction. 6 bottles (two bottles in each set) solutions do not repeat each other.
[0379] Table 7: Reaction solution in the "2+2 monochrome" method
[0380]
first bottle
second bottle
The first set
AX+CX
GX+TX
second set
AX+GX
CX+TX
third set
AX+TX
CX+GX
[0381] A complete sequencing process includes three rounds of sequenc...
Embodiment 2
[0449] Embodiment 2: Sequencing by "2+2 two-color" method
[0450] In this example, three sets of reaction solutions were prepared. There are two vials per set, each containing two nucleotide bases. The 2 nucleotide bases in each vial are labeled with two different fluorophores (so that they emit at different wavelengths) to distinguish the signals from the two nucleotide bases.
[0451] In this example, the two types of fluorophores are X and Y. For each set, two vials contained exactly all four bases of the sequencing reaction. 6 bottles (two bottles in each set) solutions are not repeated.
[0452] Table 8: Reaction solution in the "2+2 two-color" method
[0453]
first bottle
second bottle
The first set
AX+CY
GX+TY
second set
AX+GY
CX+TY
third set
AX+TY
CX+GY
[0454] A complete sequencing process includes three rounds of sequencing performed sequentially in any suitable order. Each round of sequenci...
Embodiment 3
[0464] Embodiment 3: comparative example
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com